#Loading the required package(s)
library(tidyverse)
library(car)
library(see)
library(performance)
#Reading in data and storing in object named 'rvdata' and 'wbdata'
rvdata <- read_csv("https://uoepsy.github.io/data/riverview.csv")
wbdata <- read_csv("https://uoepsy.github.io/data/wellbeing.csv")
#fit models
## riverview model
rv_mdl1 <- lm(income ~ education, data = rvdata)
## wellbeing model
wb_mdl1 <- lm(wellbeing ~ outdoor_time + social_int, data = wbdata) Assumptions and Diagnostics
Learning Objectives
At the end of this lab, you will:
- Be able to state the assumptions underlying a linear model.
- Specify the assumptions underlying a linear model with multiple predictors.
- Assess if a fitted model satisfies the assumptions of your model.
- Assess the effect of influential cases on linear model coefficients and overall model evaluations.
What You Need
Required R Packages
Remember to load all packages within a code chunk at the start of your RMarkdown file using library(). If you do not have a package and need to install, do so within the console using install.packages(" "). For further guidance on installing/updating packages, see Section C here.
For this lab, you will need to load the following package(s):
- tidyverse
- car
- see
- performance
Lab Data
You can download the data required for this lab here and here or read both datsets in via these links https://uoepsy.github.io/data/riverview.csv and https://uoepsy.github.io/data/wellbeing.csv.
Lab Overview
In the previous labs, we have fitted a number of regression models, including some with multiple predictors. In each case, we first specified the model, then visually explored the marginal distributions and relationships of variables which would be used in the analysis. Finally, we fit the model, and began to examine the fit by studying what the various parameter estimates represented, and the spread of the residuals.
But before we draw inferences using our model estimates or use our model to make predictions, we need to be satisfied that our model meets a specific set of assumptions. If these assumptions are not satisfied, the results will not hold.
In this lab, we will check the assumptions of two models that we have previously fitted - one simple linear model, and one with multiple predictors. Specifically, we will check the assumptions from the simple linear regression model we fitted using the ‘riverview’ dataset in Week 2, and the assumptions of the multiple linear regression model we fitted using the ‘wellbeing’ dataset in Week 3.
Setup
- Create a new RMarkdown file
- Load the required package(s)
- Read the riverview and wellbeing datasets into R, assigning them to objects named
rvdataandwbdatarespectively - Fit the following models:
\[ Model 1: Income = \beta_0 + \beta_1 \ Education \]
\[ Model 2: Wellbeing = \beta_0 + \beta_1 Social Interactions + \beta_2 Outdoor Time \\ \]
Exercises
Assumptions
Assumptions
You can remember the four assumptions by memorising the acronym LINE:
- Linearity: The relationship between \(y\) and \(x\) is linear.
- Independence of errors: The error terms should be independent from one another.
- Normality: The errors \(\epsilon\) are normally distributed in the population.
- Equal variances (“Homoscedasticity”): The variability of the errors \(\epsilon\) is constant across \(x\).
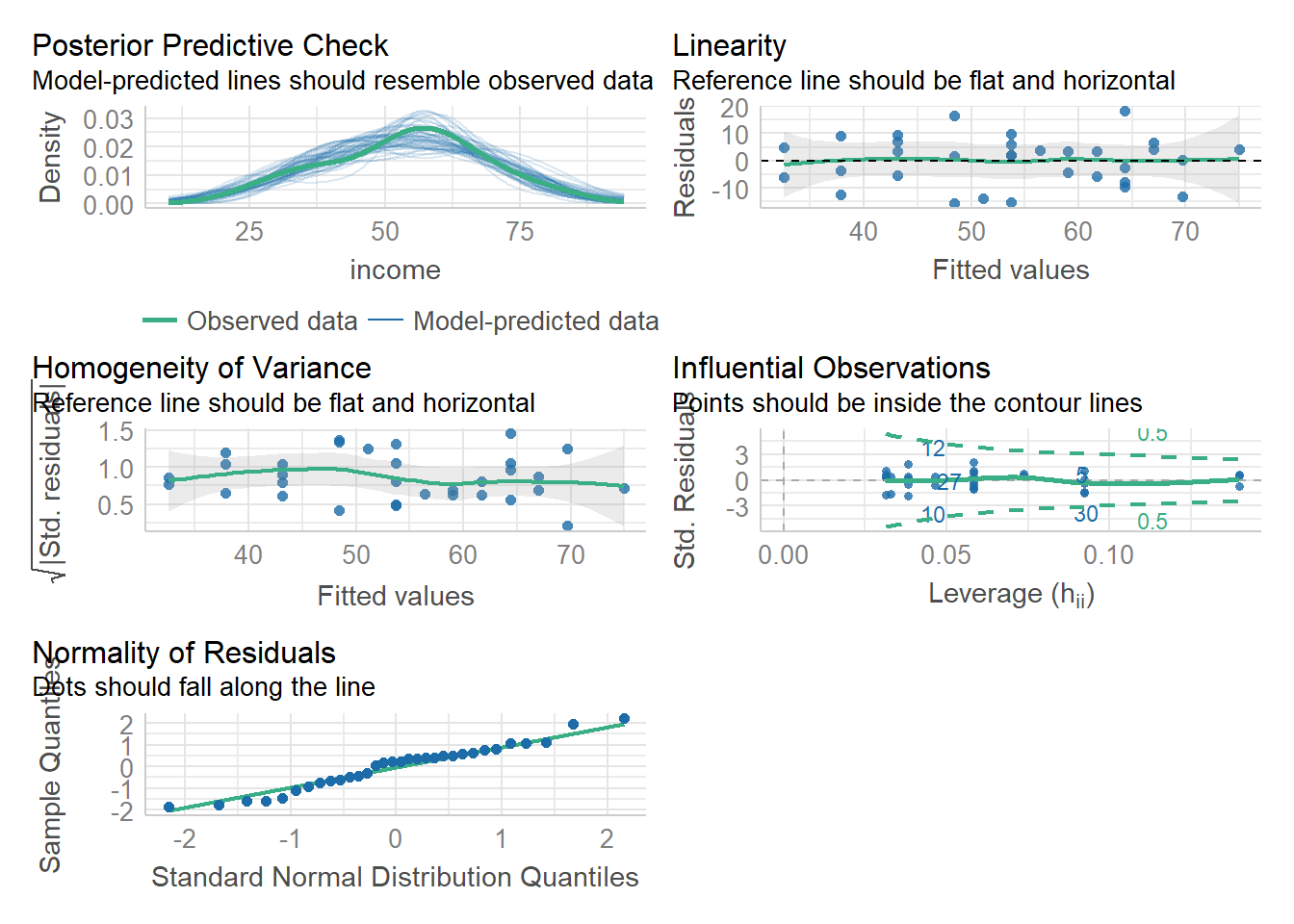
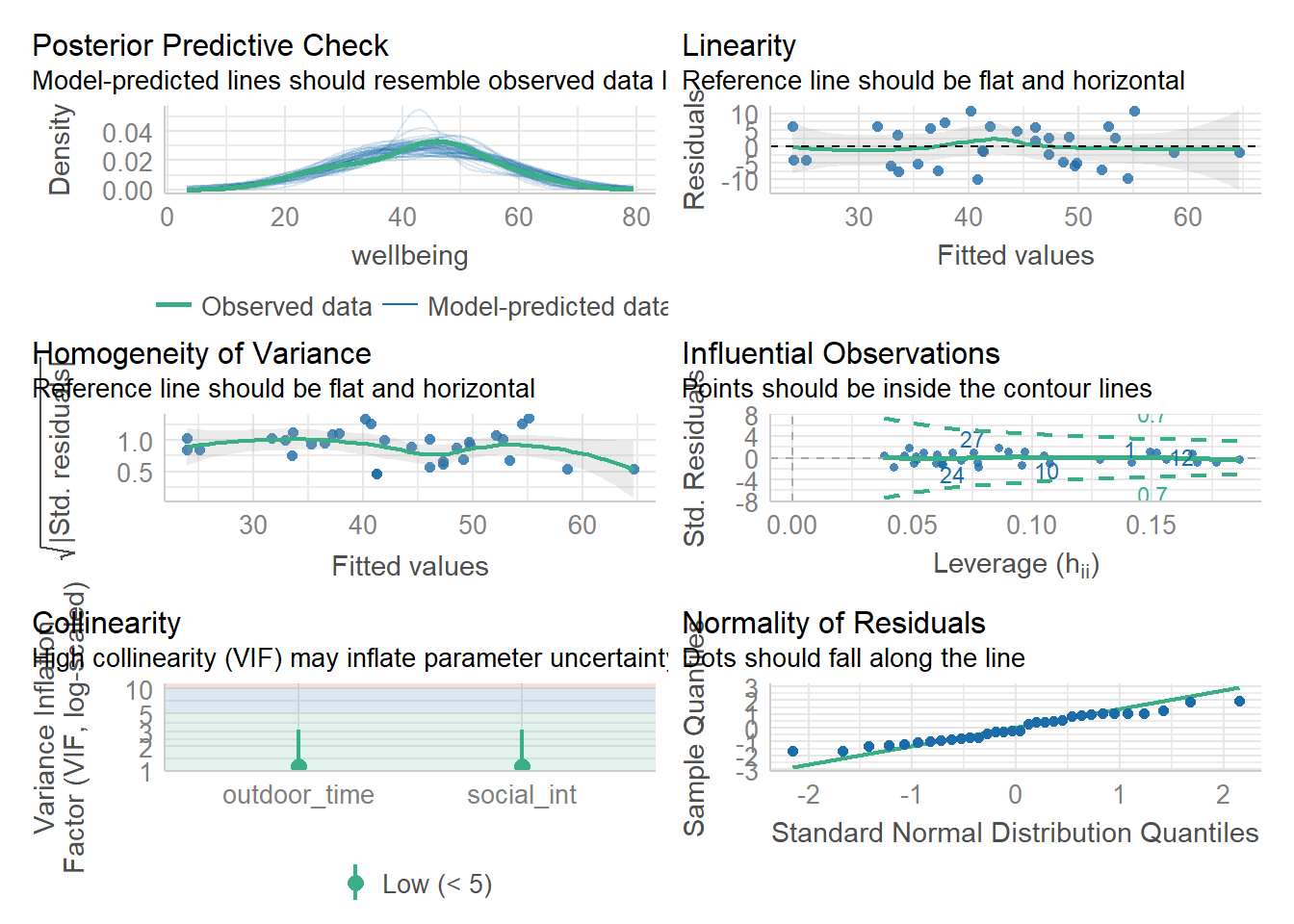
Let’s start by using check_model() for both rv_mdl1 and wb_mdl1 models - we can refer to these plots as a guide as we work through the assumptions questions of the lab.
These plots cannot be used in your reports - they are to be used as a guide only.
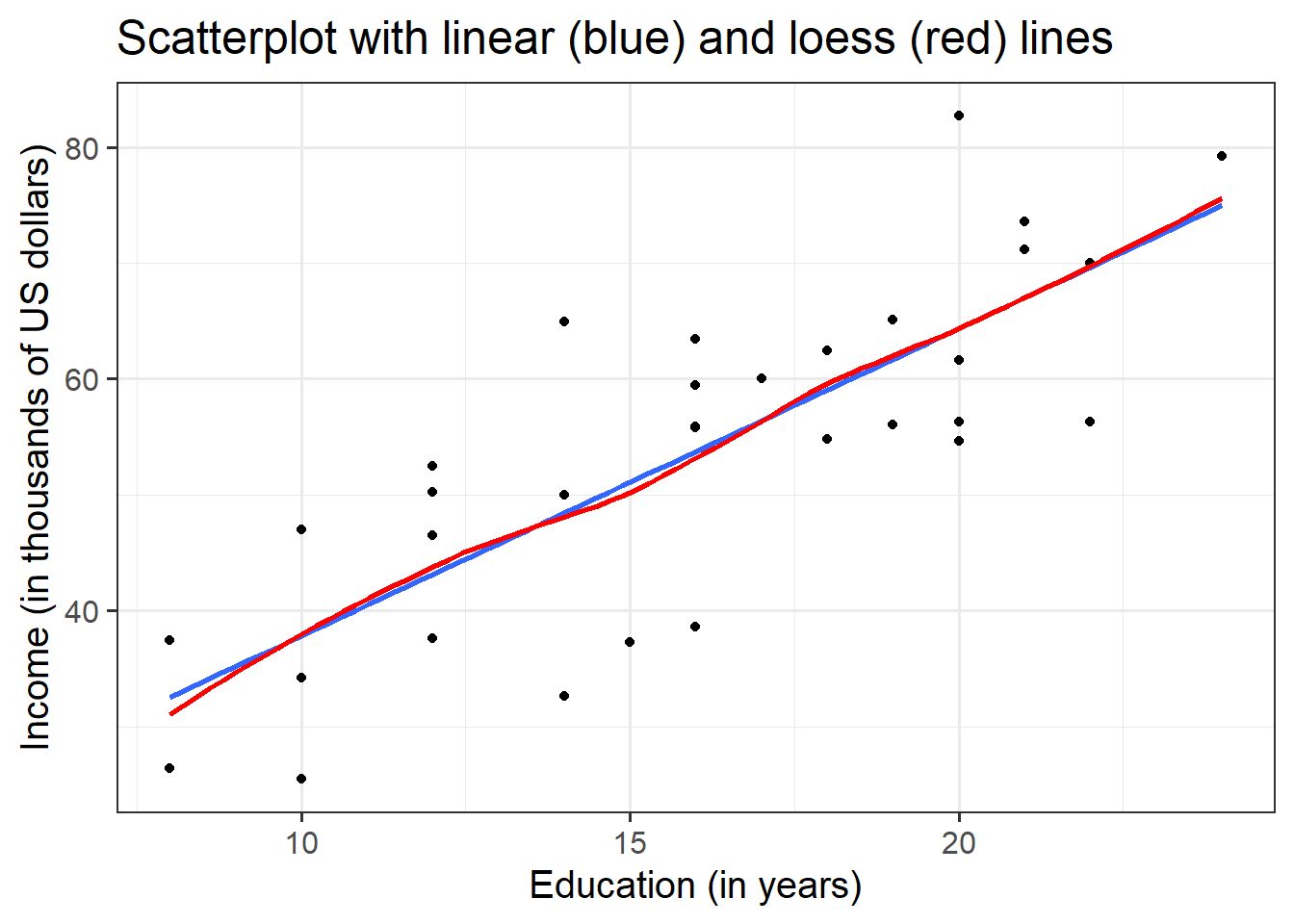
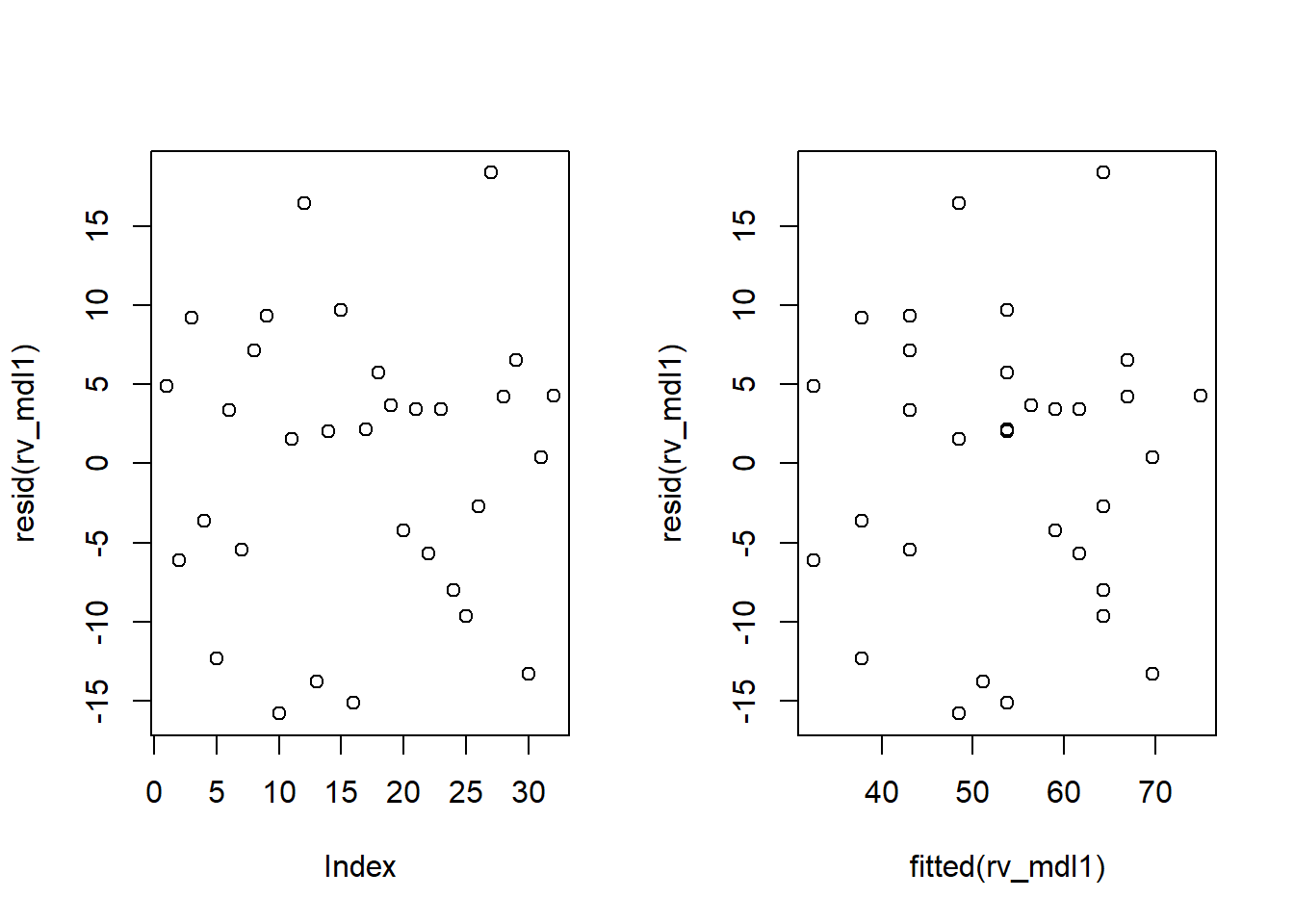
Check if the fitted model satisfies the linearity assumption for rv_mdl1.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to the plots.
Consider plotting residual vs fitted values, and/or a scatterplot with loess lines
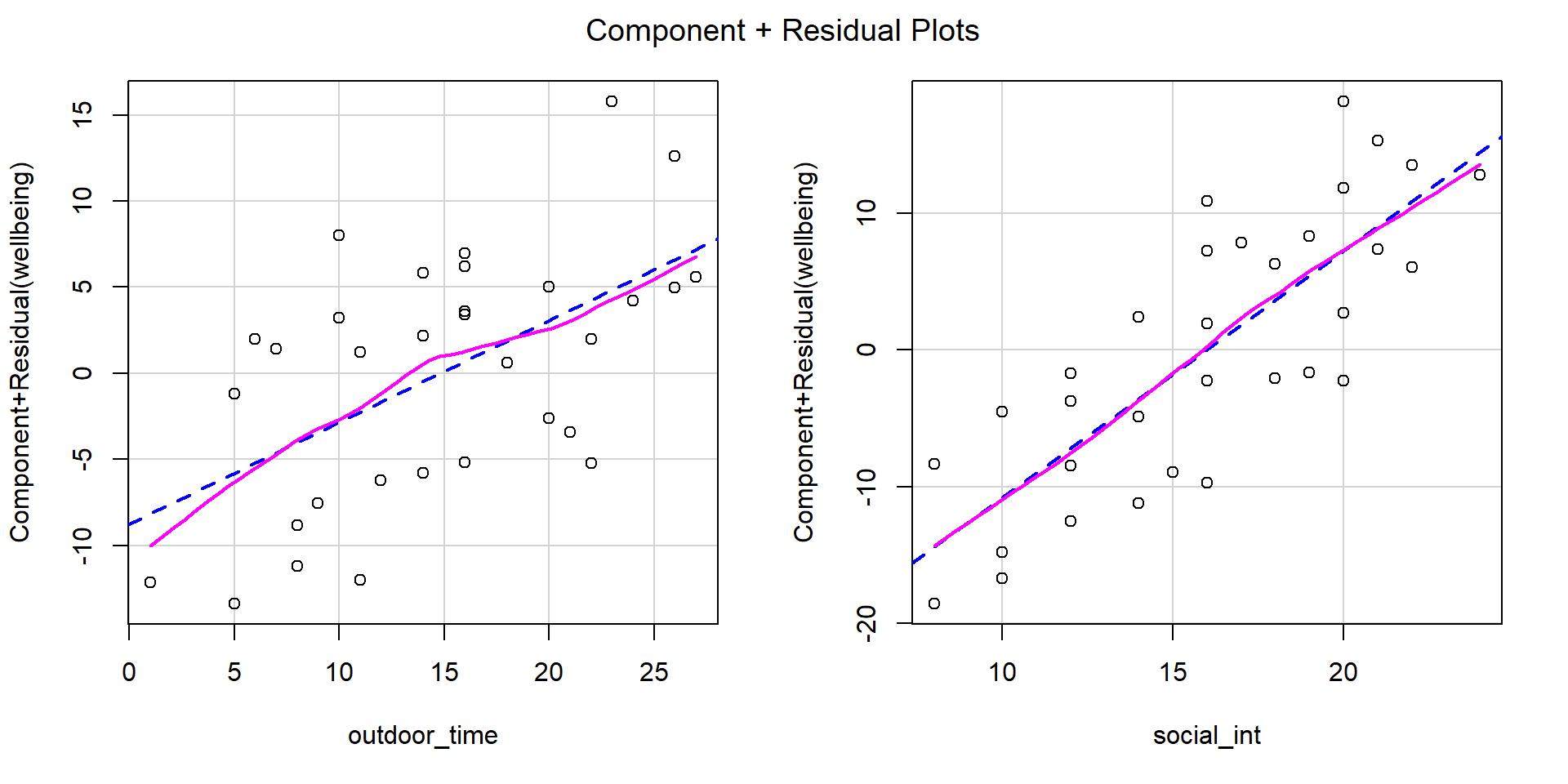
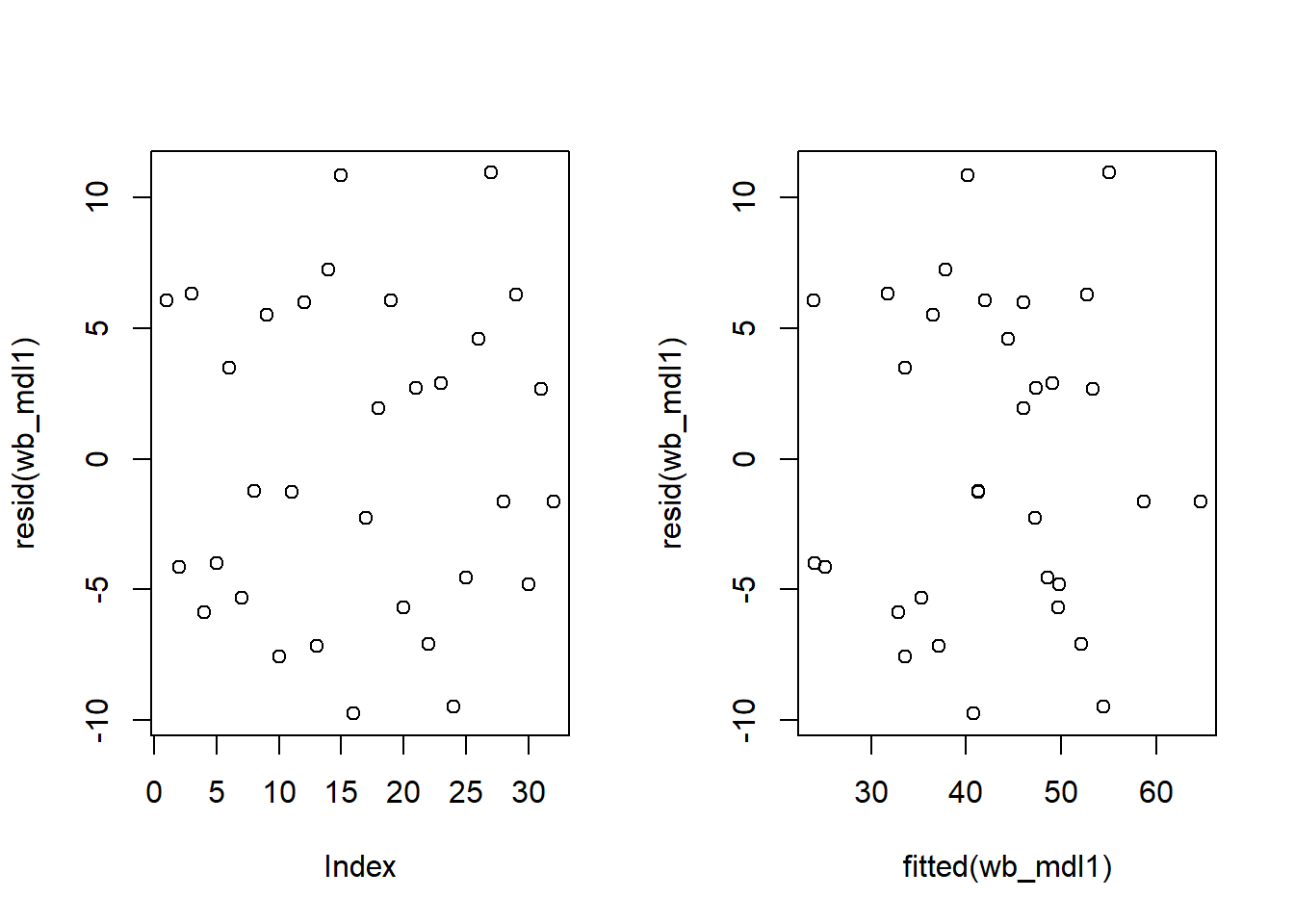
Check if the fitted model satisfies the linearity assumption for wb_mdl1.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to the plots.
When we have multiple predictors, we need to use component-residual plots (also known as partial-residual plots) to check the assumption of linearity.
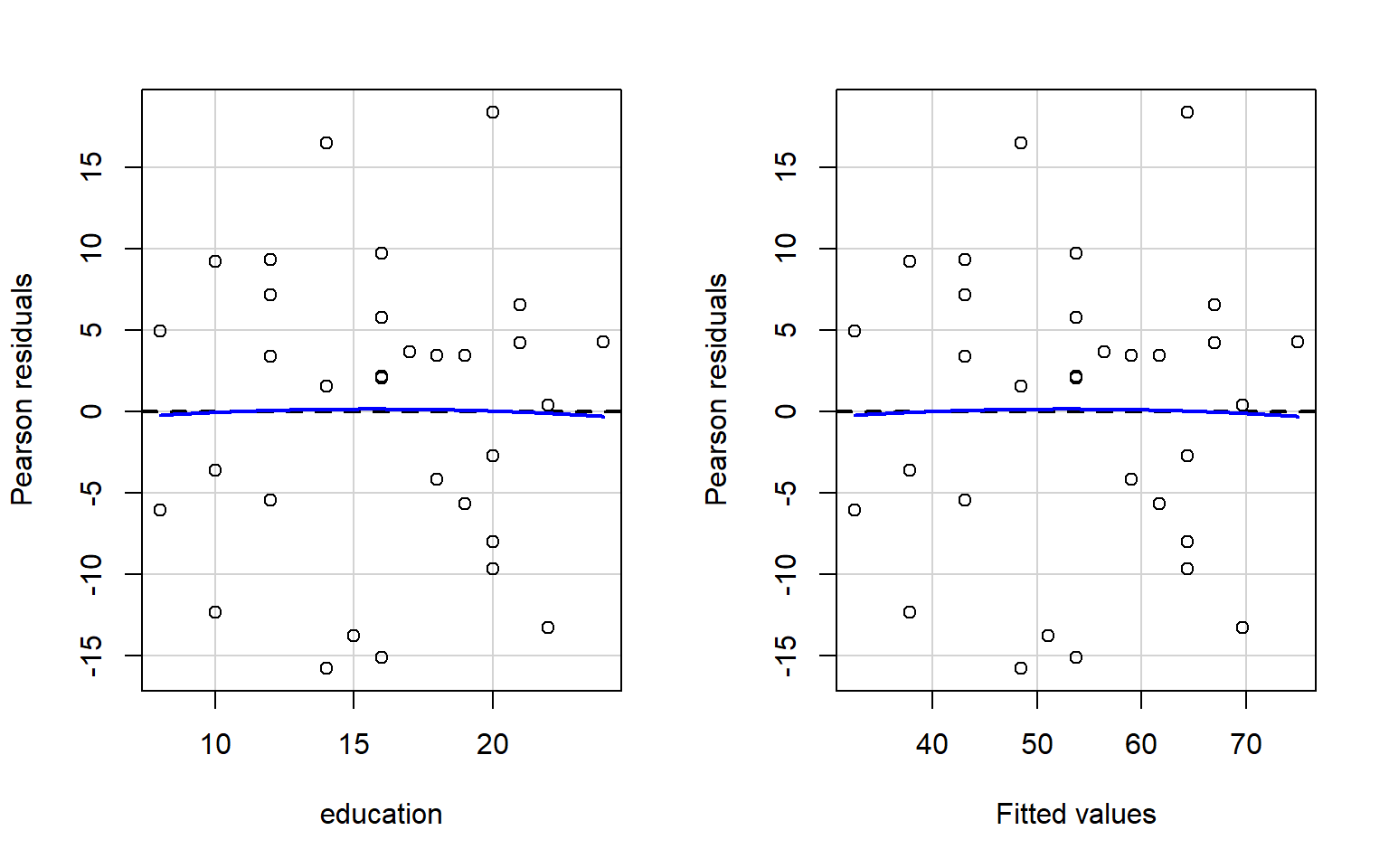
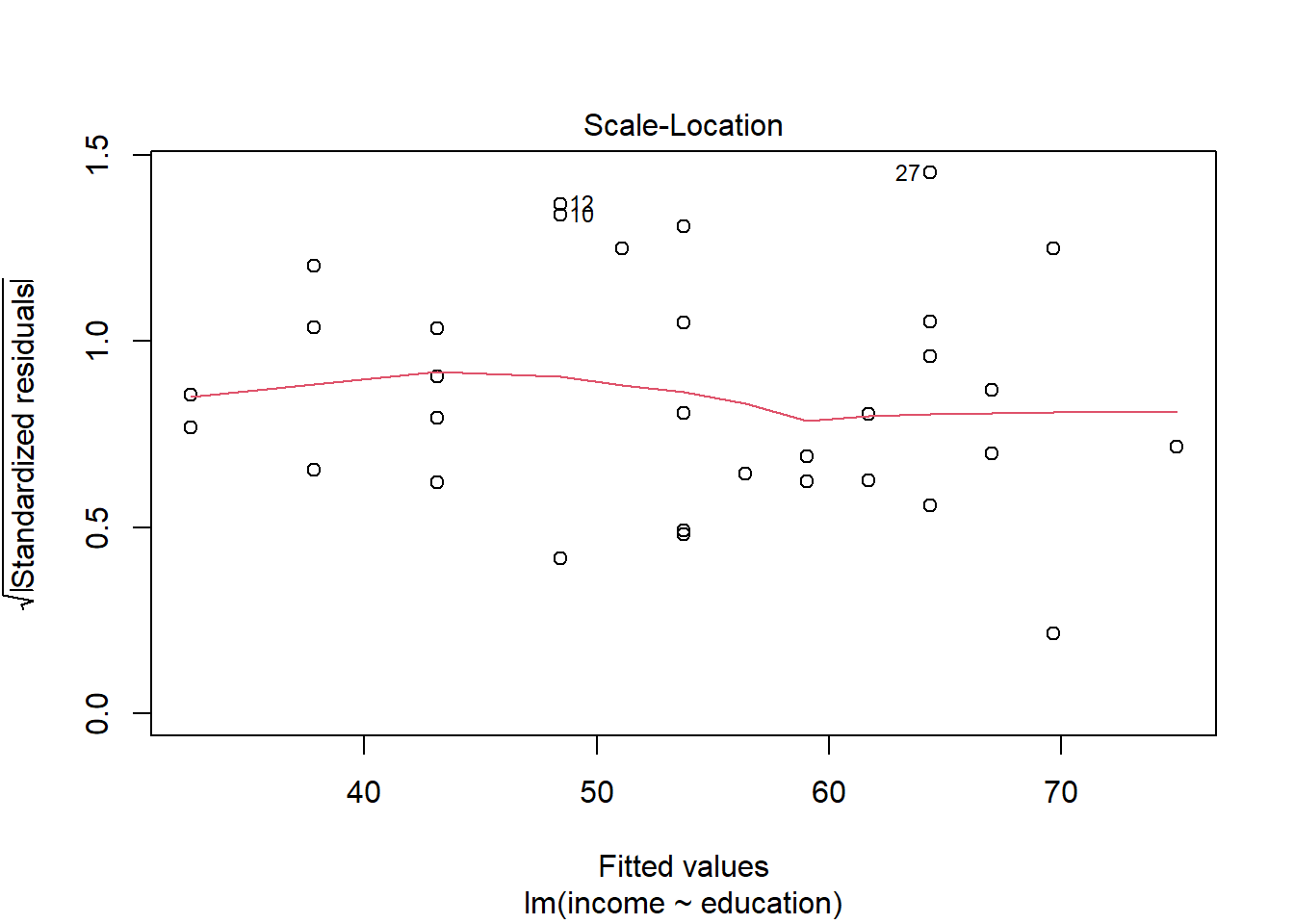
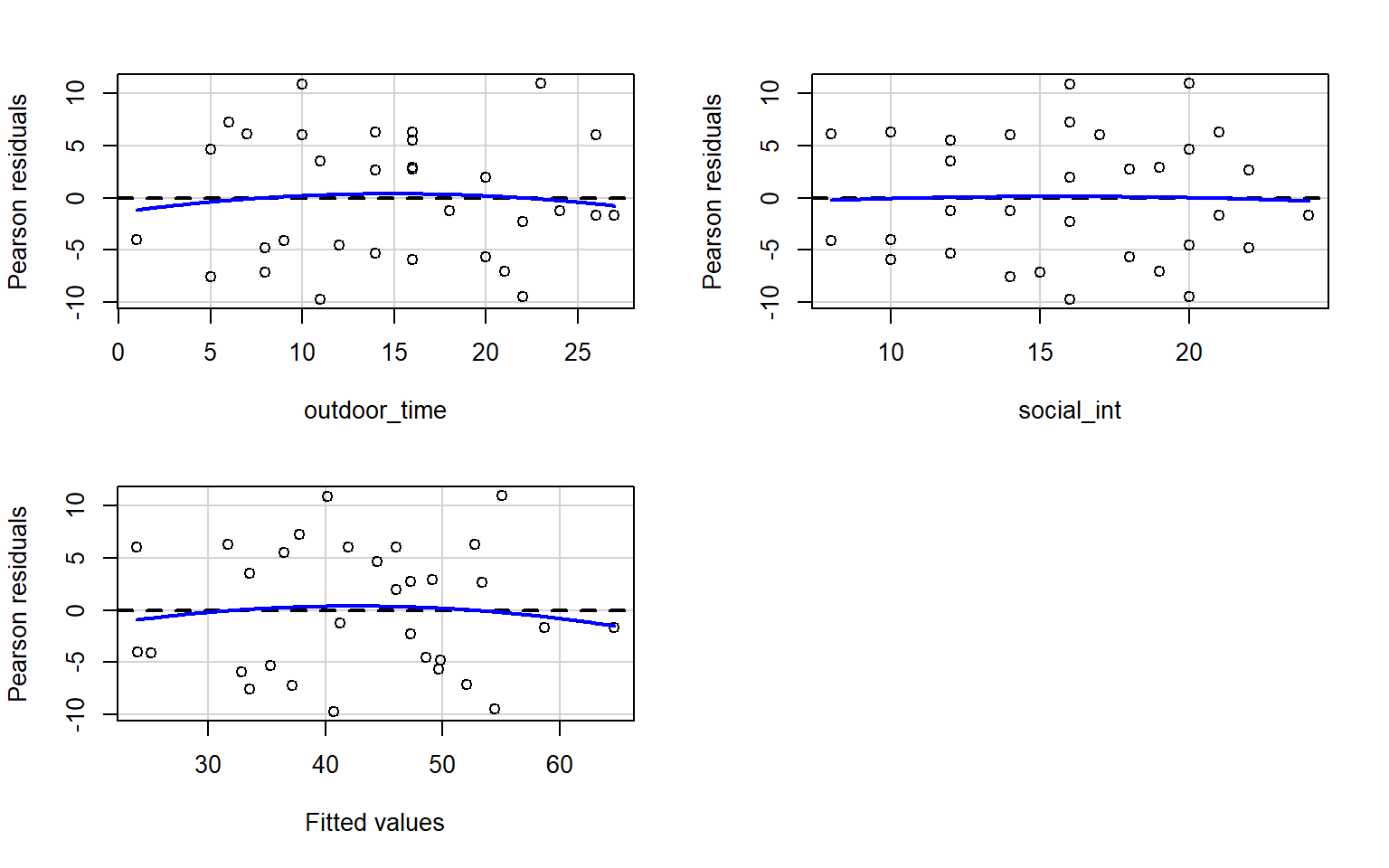
Check if the fitted models rv_mdl1 and wb_mdl1 satisfy the equal variance assumption.
Write a sentence summarising whether or not you consider the assumption to have been met for each model. Justify your answer with reference to plots.
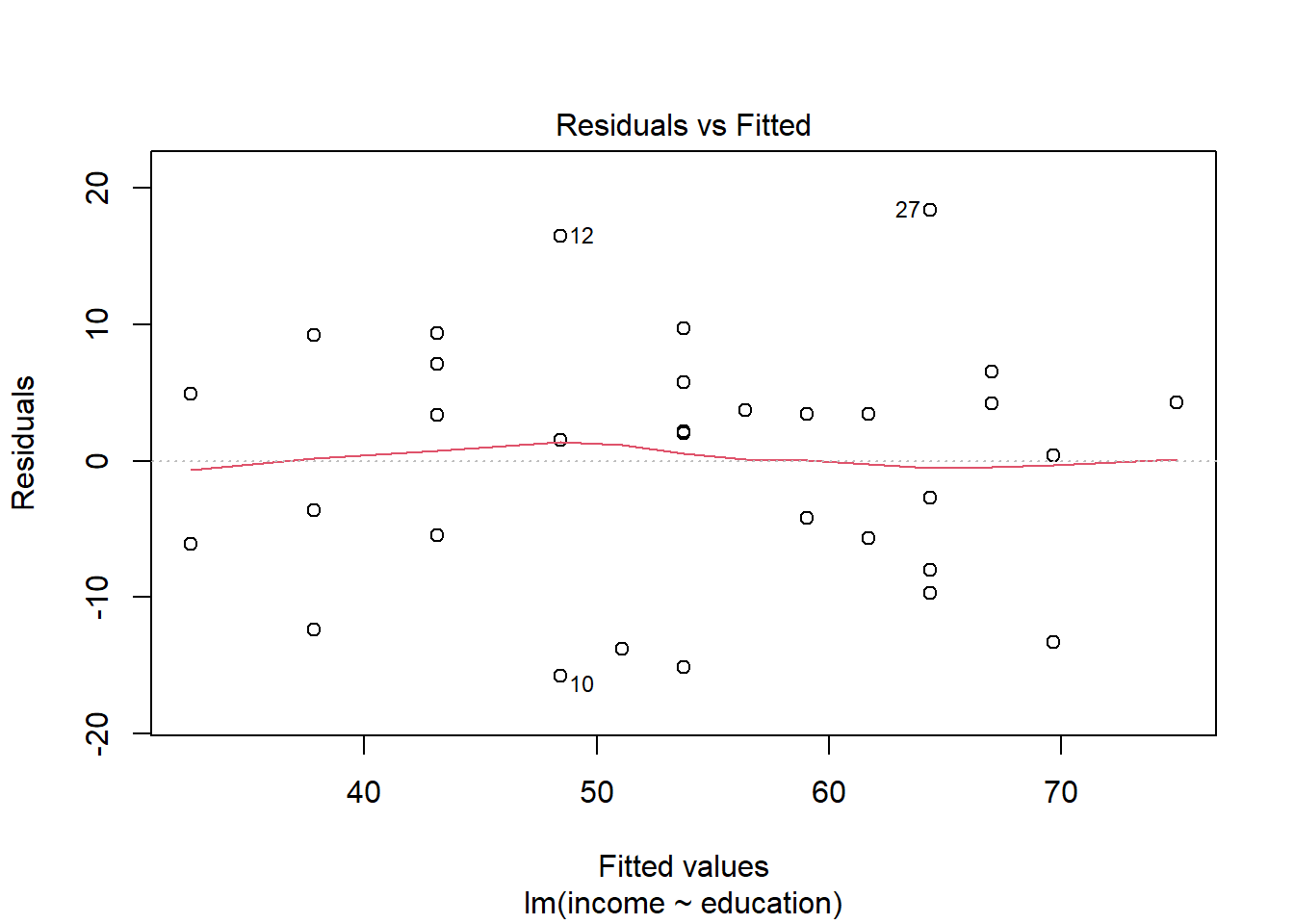
Use residualPlots() to plot residuals against the predictor.
For interpretation - the vertical spread of the residuals should roughly be the same everywhere.
For both rv_mdl1 and wb_mdl1, visually assess whether there is autocorrelation in the error terms.
Write a sentence summarising whether or not you consider the assumption of independence to have been met for each (you may have to assume certain aspects of the study design).
To get a single figure made up of 2 by 1 panels, you can use the command par(mfrow = c(1,2)). Then create the plot. Then you need to go back to a single figure made up by a single panel with the command par(mfrow = c(1,1)).
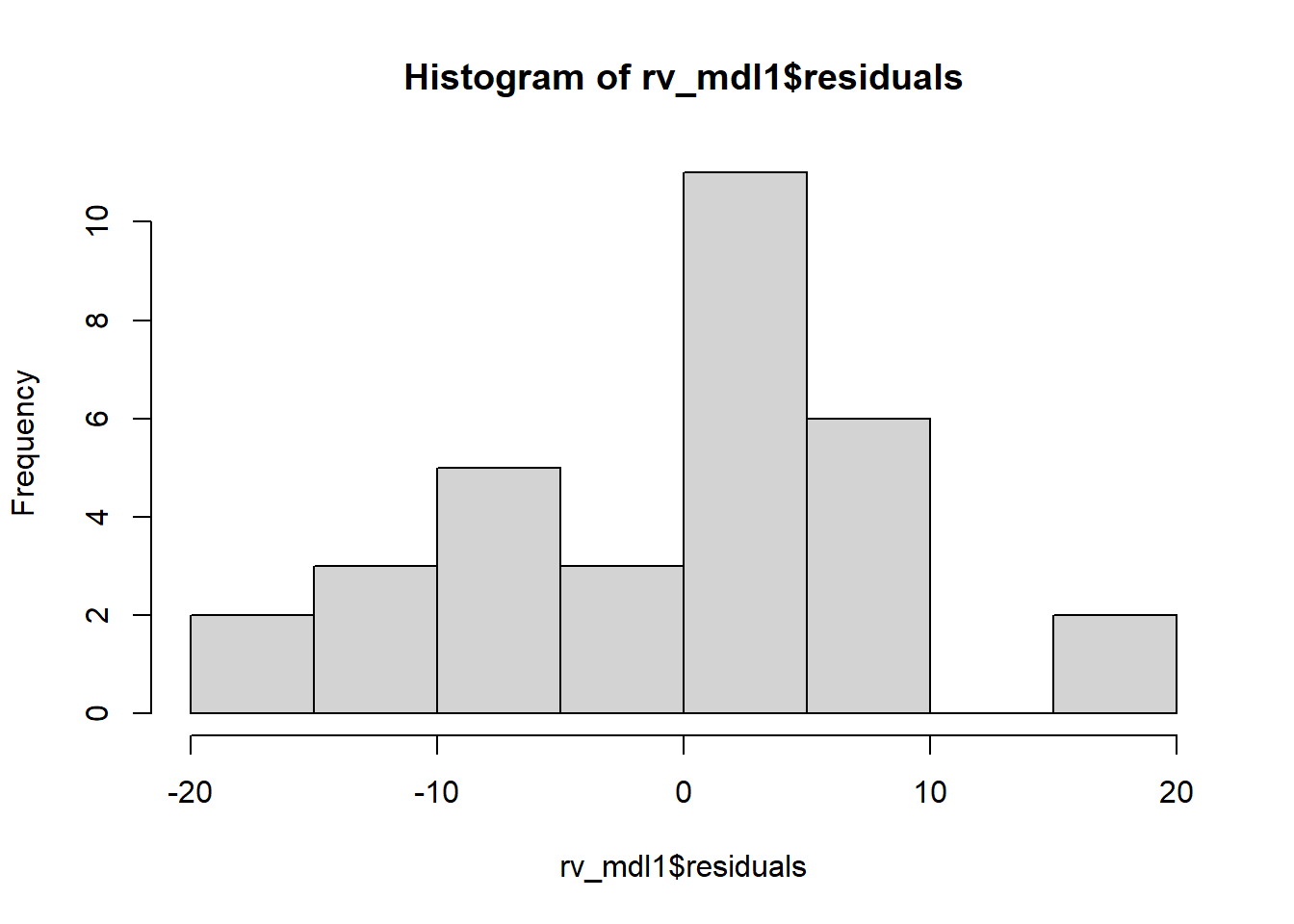
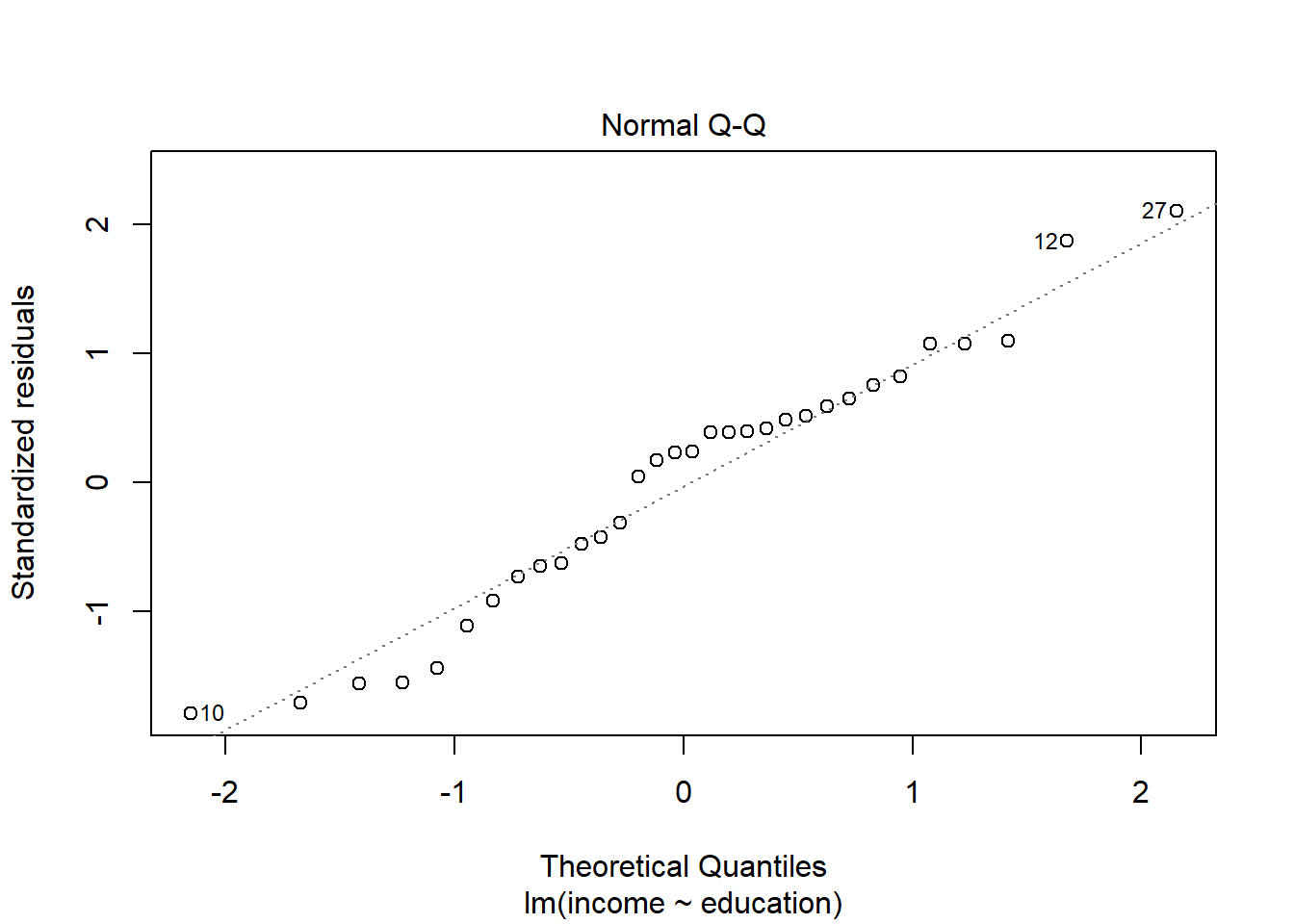
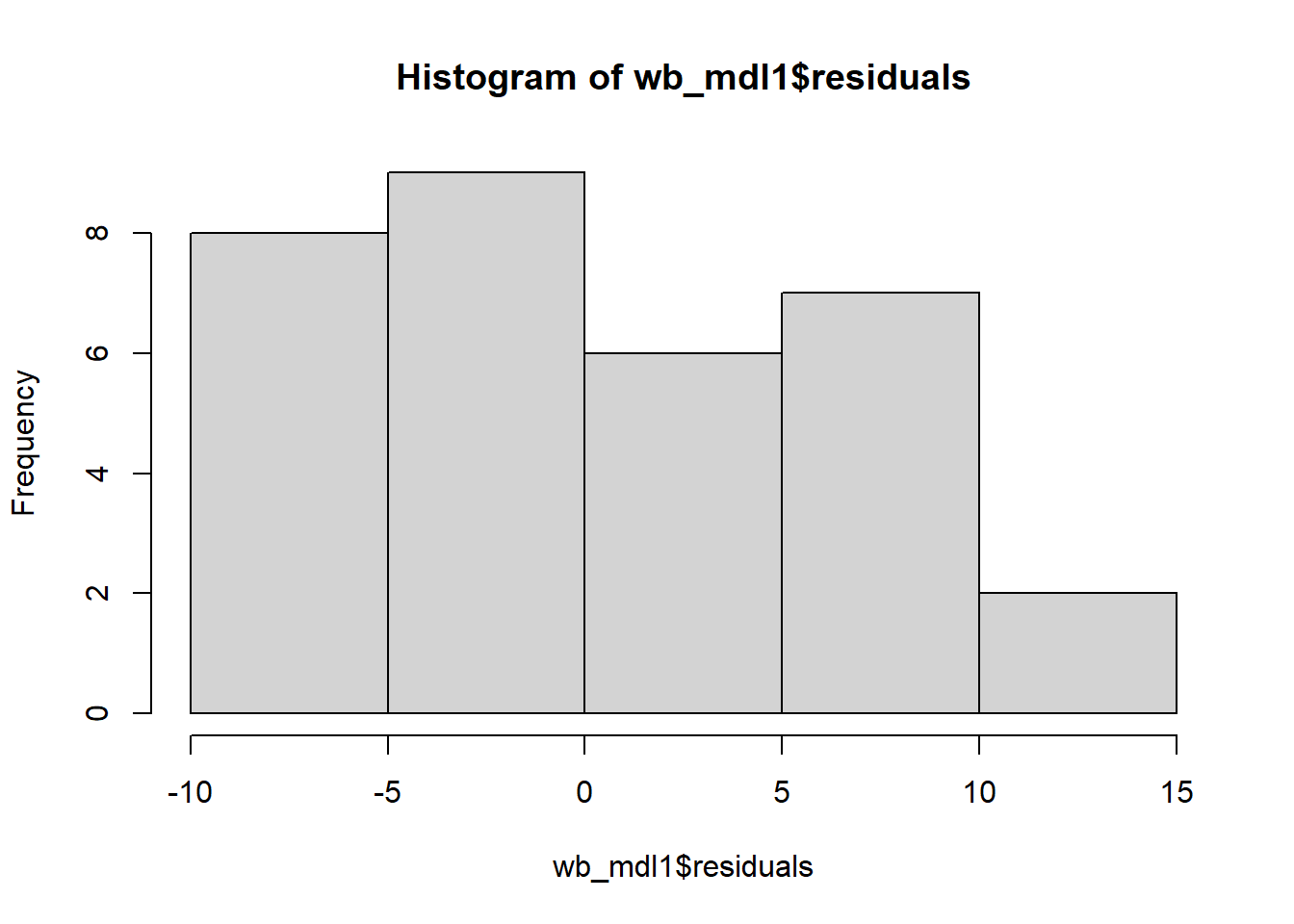
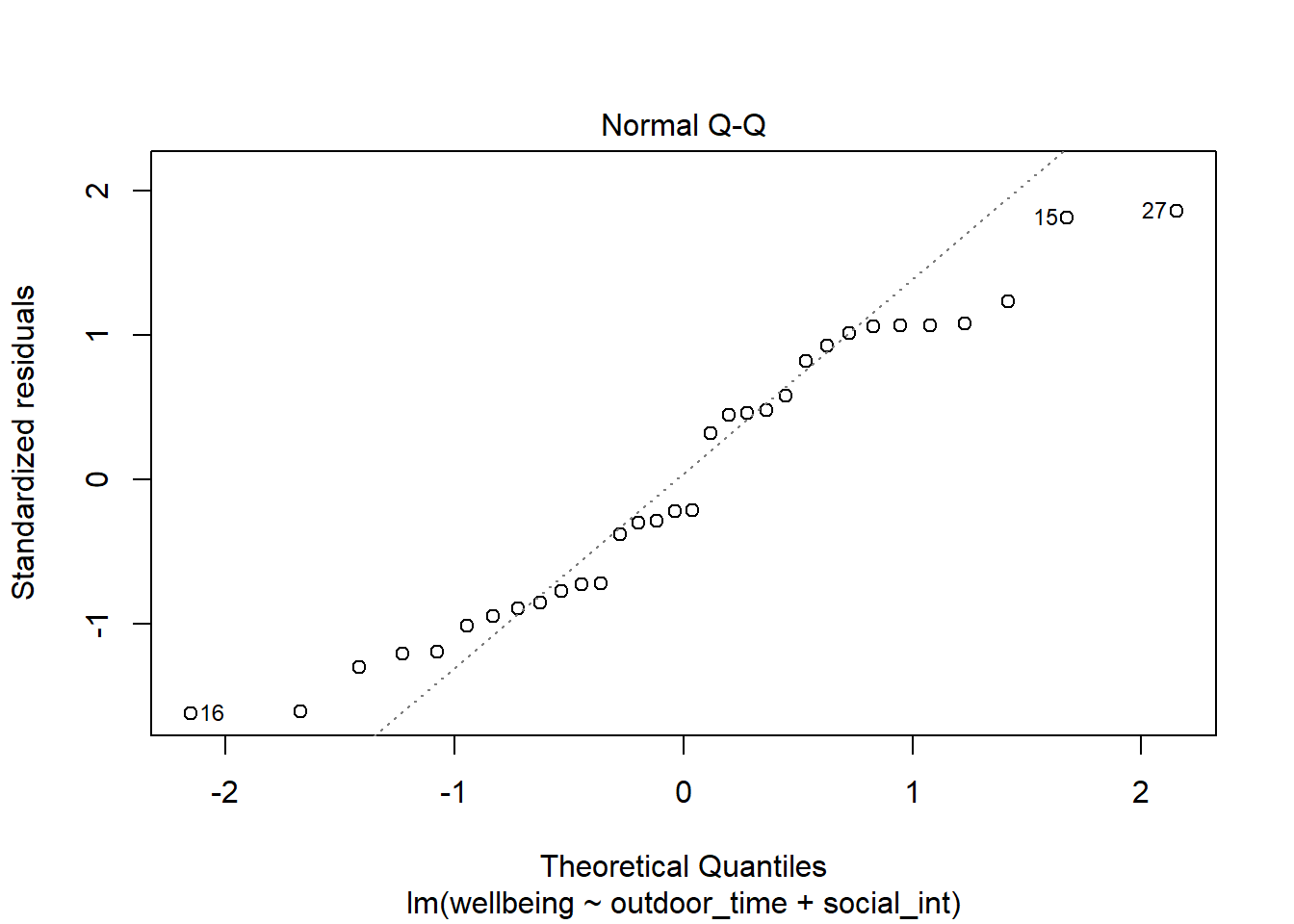
Check if the fitted models rv_mdl1 and wb_mdl1 satisfy the normality assumption.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to the plots.
We can get the QQplot from plot(model, which = ???), and you can plot the frequency distribution of the residuals via hist(model$residuals).
For interpretation - remember that departures from a linear trend in QQ plots indicate a lack of normality.
For questions 7-10, we will be working with our wb_mdl1 only. Feel free to apply the below to your rv_mdl1 too as as extra practice during revision.
Multicollinearity
For wb_mdl1, calculate the variance inflation factor (VIF) for the predictors in the model.
Write a sentence summarising whether or not you consider multicollinearity to be a problem here.
Diagnostics
Create a new tibble which contains:
- The original variables from the model (Hint, what does
wb_mdl1$modelgive you?) - The fitted values from the model \(\hat y\)
- The residuals \(\hat \epsilon\)
- The studentised residuals
- The hat values
- The Cook’s Distance values
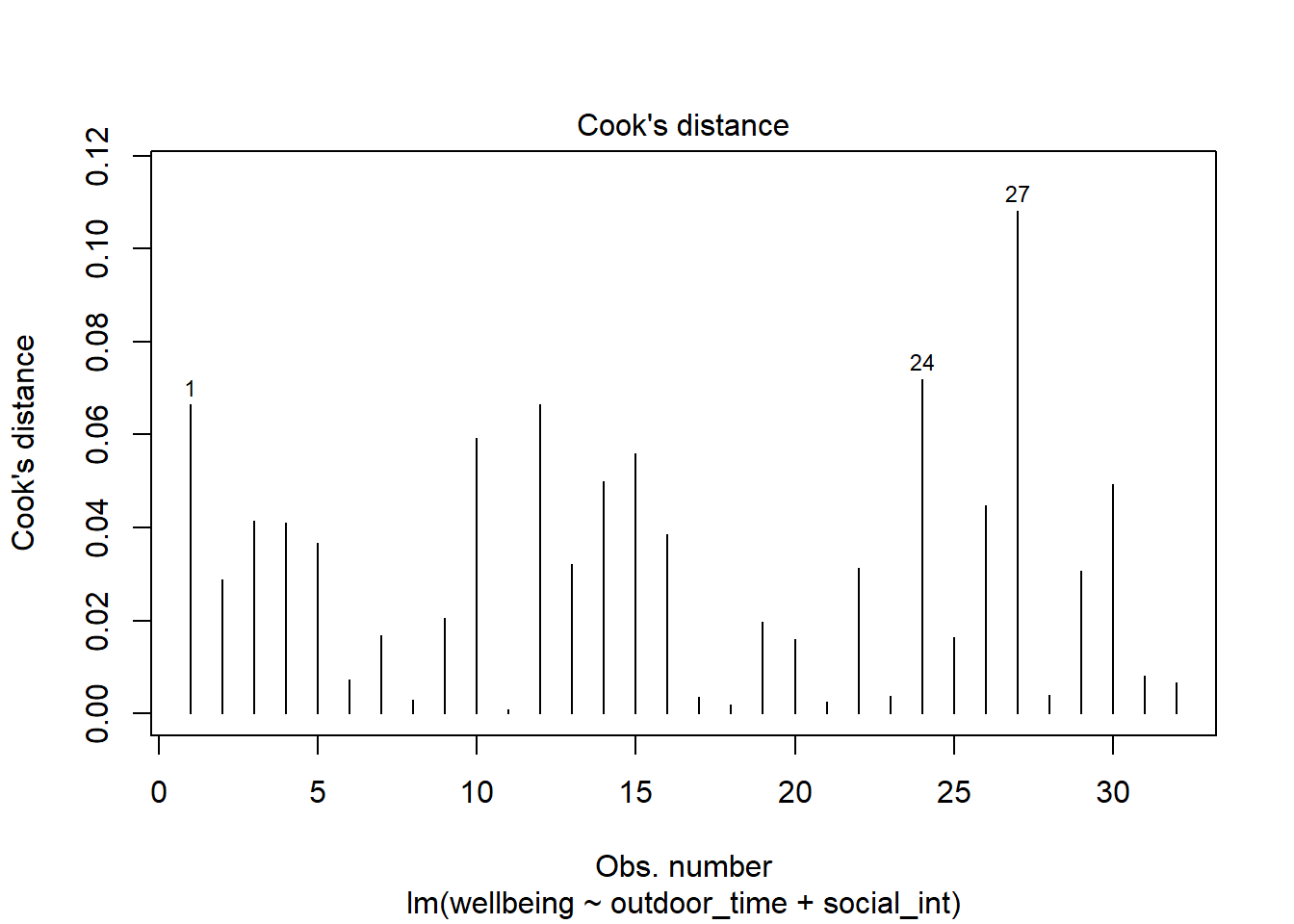
From the tibble above, comment on the following:
- Looking at the studentised residuals, are there any outliers?
- Looking at the hat values, are there any observations with high leverage?
- Looking at the Cook’s Distance values, are there any highly influential points?
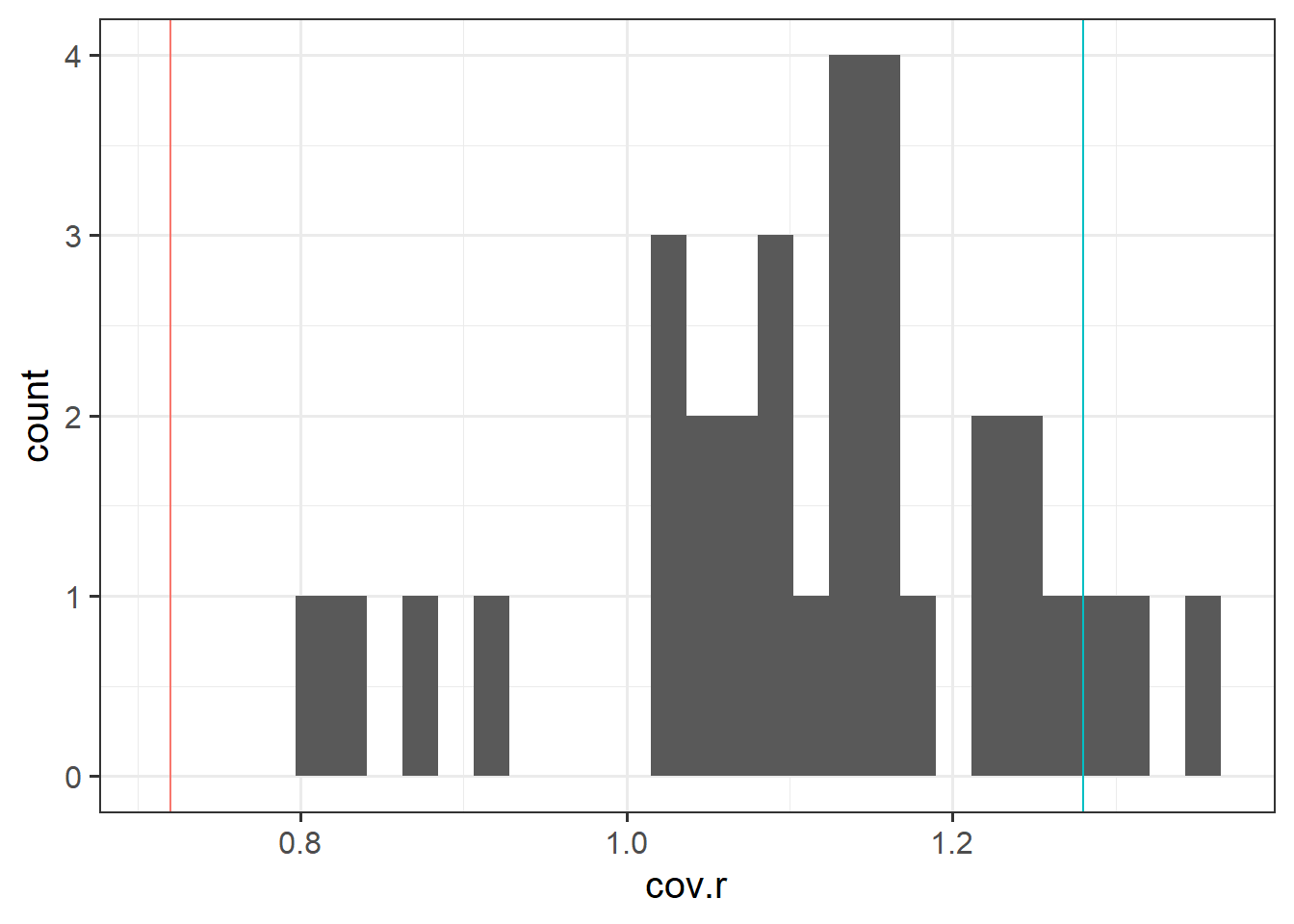
Use the function influence.measures() to extract these delete-1 measures of influence.
Try plotting the distributions of some of these measures.
The function influence.measures() returns an infl-type object. To plot this, we need to find a way to extract the actual numbers from it.
What do you think names(influence.measures(wb_mdl1)) shows you? How can we use influence.measures(wb_mdl1)$<insert name here> to extract the matrix of numbers?