Examine the dataset, and perform any necessary and appropriate data management steps.
- Convert categorical variables to factors
- Label appropriately factors to aid with your model interpretations
- If needed, provide better variable names
Note that all of these steps can be done in combination - the mutate() and factor() functions will likely be useful here.
Let’s have a look at the data to see what we’re working with:
#first look at dataset structure
str(cog)
spc_tbl_ [30 × 3] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ Diagnosis: num [1:30] 1 1 1 1 1 1 1 1 1 1 ...
$ Task : num [1:30] 1 1 1 1 1 2 2 2 2 2 ...
$ Y : num [1:30] 44 63 76 72 45 70 51 82 66 56 ...
- attr(*, "spec")=
.. cols(
.. Diagnosis = col_double(),
.. Task = col_double(),
.. Y = col_double()
.. )
- attr(*, "problems")=<externalptr>
#now lets look at top 6 rows (or the head) of the dataset
head(cog)
# A tibble: 6 × 3
Diagnosis Task Y
<dbl> <dbl> <dbl>
1 1 1 44
2 1 1 63
3 1 1 76
4 1 1 72
5 1 1 45
6 1 2 70
The columns Diagnosis and Task should be coded into factors with better labels, as currently, without making reference to the codebook, it is not clear what “1” and “2” represent. It is also unclear what the Y column represents - this should be renamed.
#We can make all of the changes noted above in one (long) command.
#First we can use the function `factor()` by specifying the current levels and what labels each level should map to.
#We can also simply rename the Y column to score.
cog <- cog %>%
mutate(
Diagnosis = factor(Diagnosis,
levels = c(1, 2, 3),
labels = c('Amnesic', 'Huntingtons', 'Control')),
Task = factor(Task,
levels = c(1, 2),
labels = c('Grammar', 'Recognition'))) %>%
rename(Score = Y)
#Use head() function to check renaming
head(cog)
# A tibble: 6 × 3
Diagnosis Task Score
<fct> <fct> <dbl>
1 Amnesic Grammar 44
2 Amnesic Grammar 63
3 Amnesic Grammar 76
4 Amnesic Grammar 72
5 Amnesic Grammar 45
6 Amnesic Recognition 70
Choose appropriate reference levels for the Diagnosis and Task variables.
The Diagnosis factor has a group coded ‘Control’ which lends itself naturally to be the reference category.
cog$Diagnosis <- relevel(cog$Diagnosis, 'Control')
levels(cog$Diagnosis)
[1] "Control" "Amnesic" "Huntingtons"
There is no natural reference category for the Task factor, so we will leave it unaltered. However, if you are of a different opinion, please note that there is no absolute correct answer. As long as you interpret the model correctly, you will reach to the same conclusions as someone that has chosen a different baseline category.
Before stating the model, you first need to define the dummy variables for Diagnosis:
\[
D_\text{Amnesic} = \begin{cases}
1 & \text{if Diagnosis is Amnesic} \\
0 & \text{otherwise}
\end{cases}
\quad
D_\text{Huntingtons} = \begin{cases}
1 & \text{if Diagnosis is Huntingtons} \\
0 & \text{otherwise}
\end{cases}
\quad
(\text{Control is base level})
\]
And for Task:
\[
T_\text{Recognition} = \begin{cases}
1 & \text{if Task is Recognition} \\
0 & \text{otherwise}
\end{cases}
\quad
(\text{Grammar is base level})
\] The model becomes:
\[
\begin{aligned}
Score &= \beta_0 \\
&+ \beta_1 D_\text{Amnesic} + \beta_2 D_\text{Huntingtons} \\
&+ \beta_3 T_\text{Recognition} \\
&+ \beta_4 (D_\text{Amnesic} * T_\text{Recognition}) + \beta_5 (D_\text{Huntingtons} * T_\text{Recognition}) \\
&+ \epsilon
\end{aligned}
\] Effects will be considered statistically significant at \(\alpha=.05\)
Our hypotheses are:
\(H_0: \beta_5 = 0\)
The difference in performance between explicit and implicit memory tasks does not significantly differ between patients with Huntingtons in comparison to Controls.
\(H_1: \beta_5 \neq 0\)
The difference in performance between explicit and implicit memory tasks does significantly differ between patients with Huntingtons in comparison to Controls.
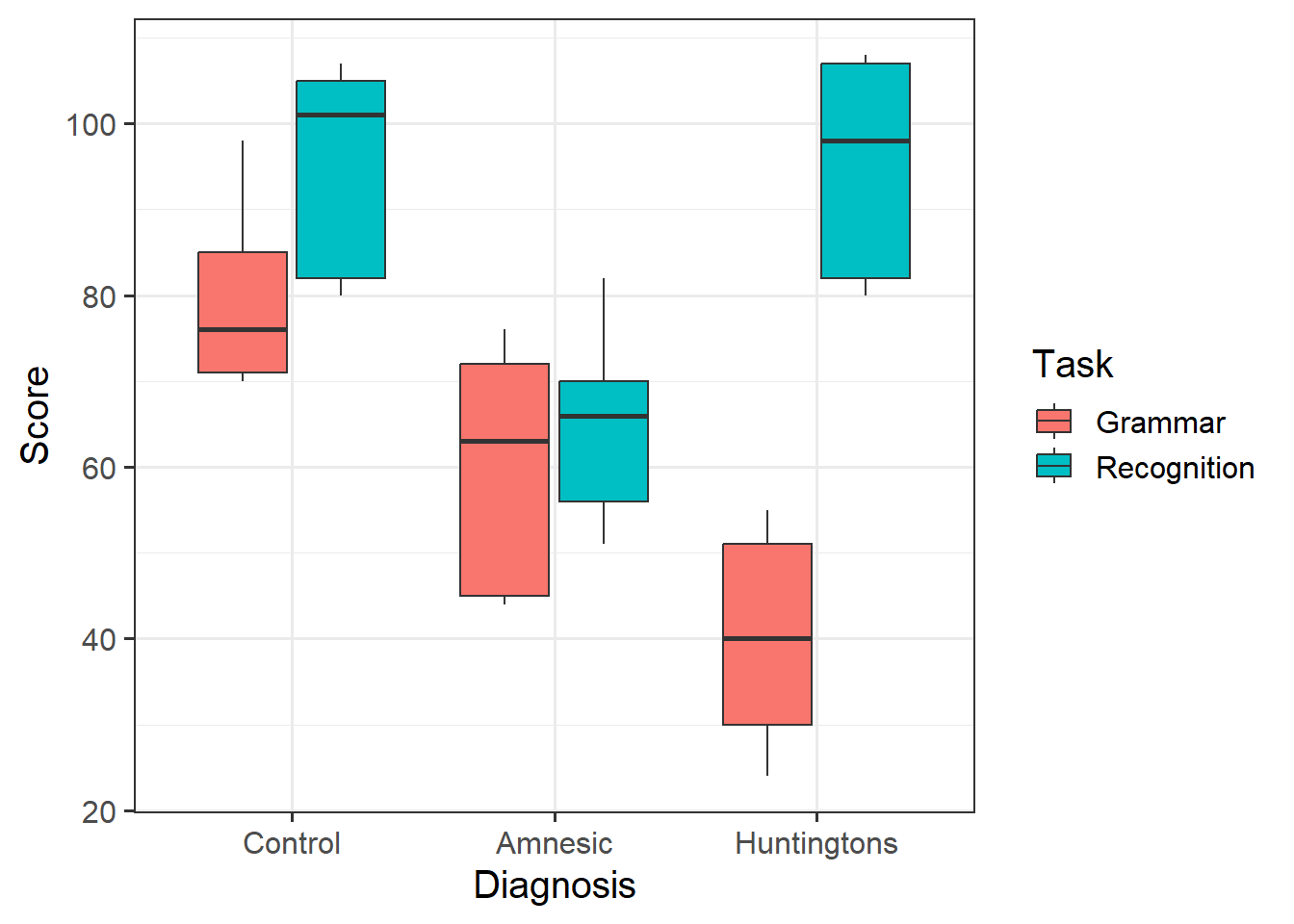
Provide a table of descriptive statistics and visualise your data.
Remember to interpret your plot in the context of the study.
- For your table of descriptive statistics, both the
group_by() and summarise() functions will come in handy here.
- Recall that when visualising categorical variables,
geom_boxplot() may be most appropriate to use.
First, lets look at our descriptive statistics and present in a well formatted table:
Table 1: Descriptive Statistics
| Diagnosis |
Task |
Mean |
SD |
Min |
Max |
| Control |
Grammar |
80 |
11.68 |
70 |
98 |
| Control |
Recognition |
95 |
12.98 |
80 |
107 |
| Amnesic |
Grammar |
60 |
14.92 |
44 |
76 |
| Amnesic |
Recognition |
65 |
12.17 |
51 |
82 |
| Huntingtons |
Grammar |
40 |
13.25 |
24 |
55 |
| Huntingtons |
Recognition |
95 |
13.38 |
80 |
108 |
Next, look at associations among variables of interest:
Participants with Amnesia do not appear to differ in Score for Recognition or Grammar tasks. In comparison to Controls, Amnesic patients score lower on both tasks, but not considerably so.
Participants with Huntingtons do differ in Score for Recognition and Grammar tasks, with higher scores on Recognition tasks. In comparison to Controls, Huntingtons patients score similarly on Recognition tasks, but considerably lower on Grammar tasks.
Fit the specified model using lm(), and store the model in an object named “cog_mdl”.
Fortunately, R computes the dummy variables for us! Thus. each row in the summary() output of the model will correspond to one of the estimated \(\beta\)’s in the equation above.
Recall your table of descriptive statistics - map each coefficient from the summary() output from “cog_mdl” to the group means.
Interpret your coefficients in the context of the study.
Recall that we can obtain our parameter estimates using various functions such as summary(),coef(), coefficients(), etc.
(Intercept) DiagnosisAmnesic
80 -20
DiagnosisHuntingtons TaskRecognition
-40 15
DiagnosisAmnesic:TaskRecognition DiagnosisHuntingtons:TaskRecognition
-10 40
-
\(\beta_0\) =
(Intercept) = 80
- The intercept, or predicted scores for those in the Control diagnosis condition on the Grammar task.
-
\(\beta_1\) =
DiagnosisAmnesic = -20
- The difference in scores between Amnesic and Control conditions on the Grammar task
- On the Grammar task, individuals with Amnesia scored 20 points lower than Control participants.
-
\(\beta_2\) =
DiagnosisHuntingtons = -40
- The difference in score between Huntingtons and Control conditions on the Grammar task
- On the Grammar task, individuals with Huntingtons scored 40 points lower than Control participants.
-
\(\beta_3\)
TaskRecognition = 15
- The difference in score between individuals in the Control diagnosis condition completing Recognition and Grammar tasks.
- Control participants scored 15 points higher when completing Recognition tasks in comparison to Grammar tasks.
-
\(\beta_4\)
DiagnosisAmnesic:TaskRecognition = -10
- The difference between score in Amnesic and Control diagnosis conditions between Recognition and Grammar tasks.
- The difference between Grammar and Recognition tasks is 10 points lower in the Amnesiac vs Control diagnosis conditions.
-
\(\beta_5\)
DiagnosisHuntingtons:TaskRecognition = 40
- The difference between score in Huntingtons and Control diagnosis conditions between Recognition and Grammar tasks.
- The difference between Grammar and Recognition tasks is 40 points higher in the Huntingtons vs Control diagnosis conditions.
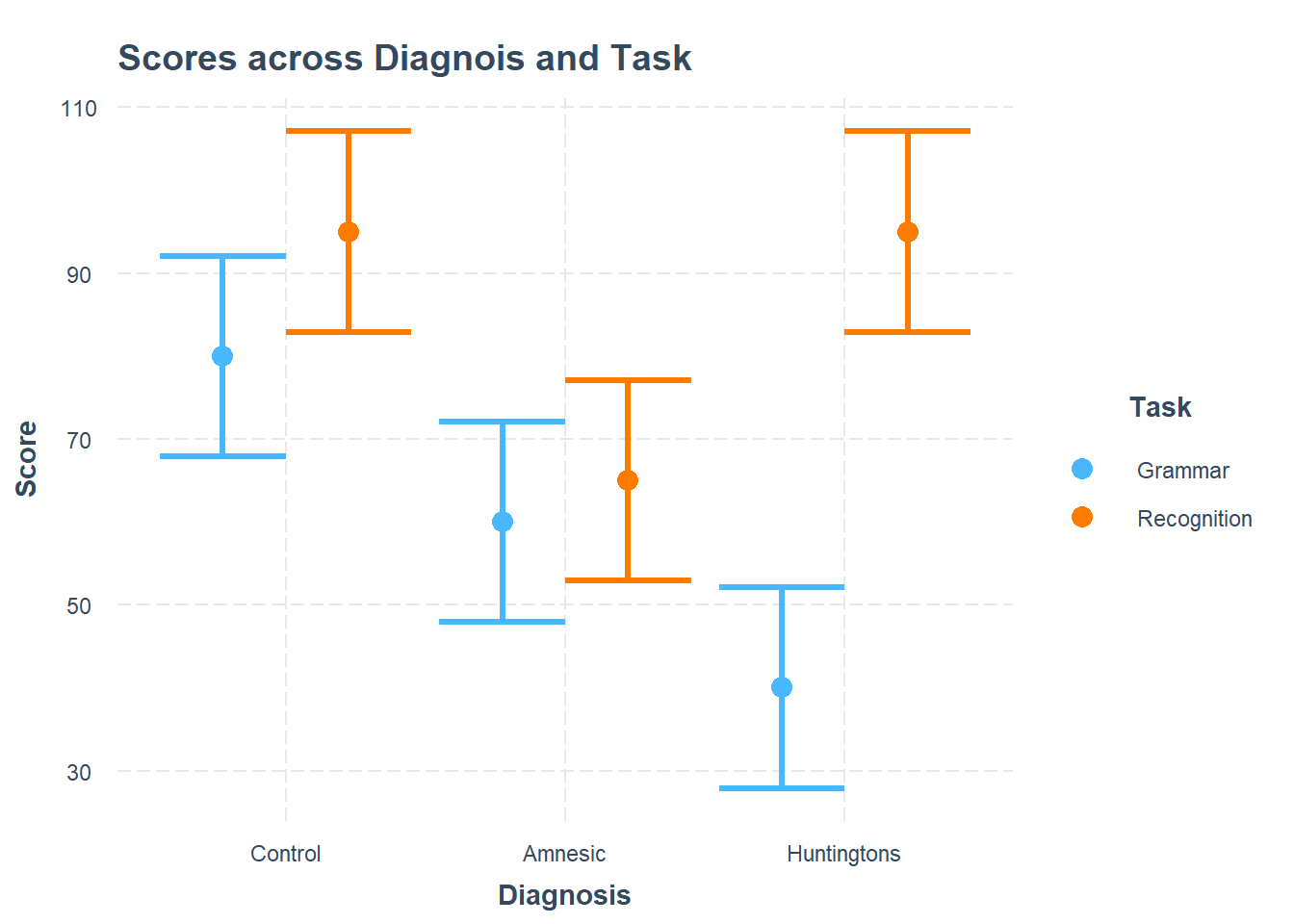
Using the cat_plot() function from the interactions package, visualise the interaction effects from your model.
Try to summarise the interaction effects in a few short and concise sentences.
plt_cog_mdl <- cat_plot(model = cog_mdl,
pred = Diagnosis,
modx = Task,
main.title = "Scores across Diagnois and Task",
x.label = "Diagnosis",
y.label = "Score",
legend.main = "Task")
plt_cog_mdl
The effect of Task on Scores does appear to vary depending on Diagnosis.
The difference in score between recognition and grammar tasks for Huntingtons patients is larger than the difference in score between recognition and grammar tasks for the Control patients.
The difference in score between recognition and grammar tasks for Amnesic patients however does not appear to be very different (given the overlapping intervals) than the difference in score between recognition and grammar tasks for the Control patients.
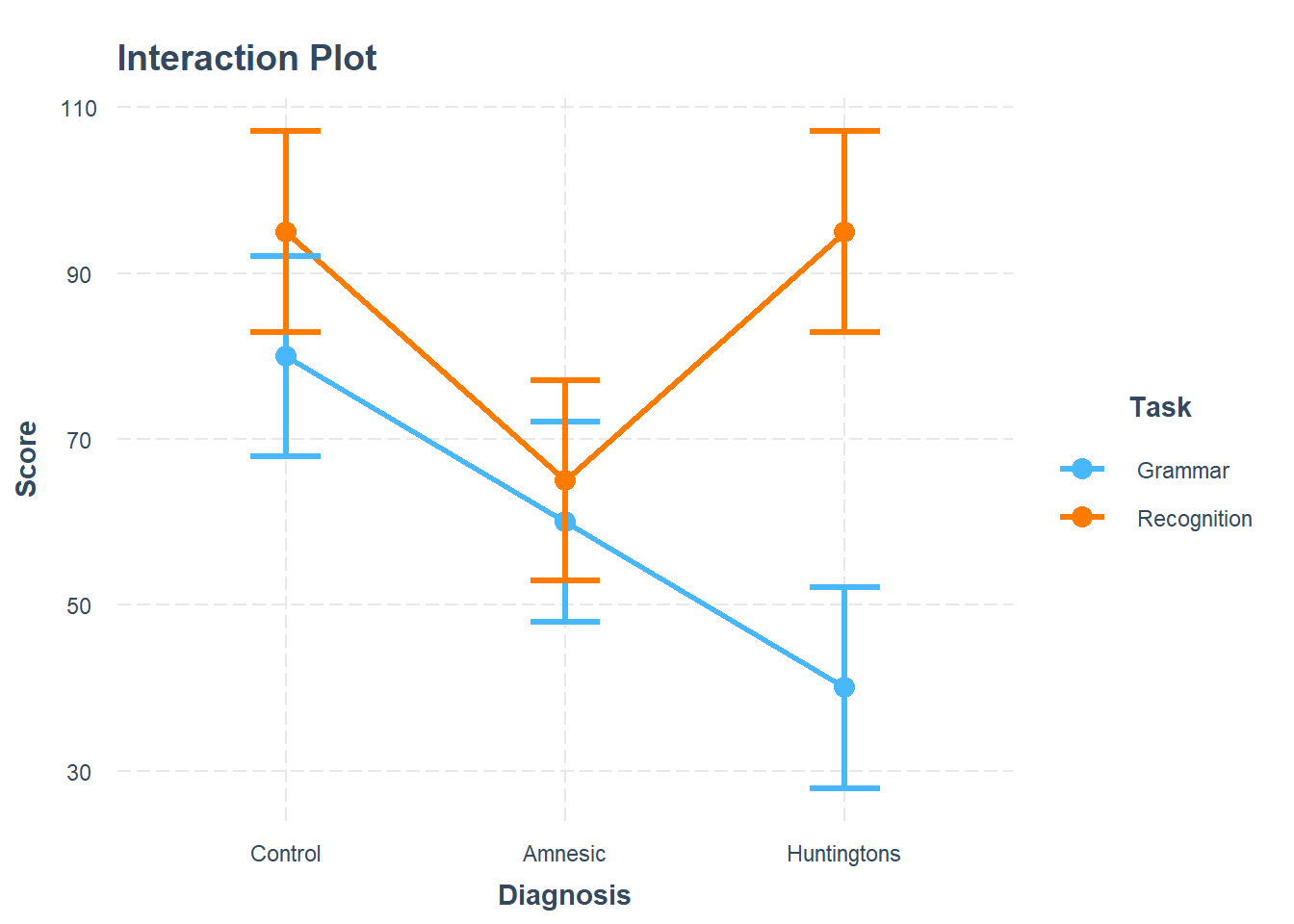
If you imagine connecting the dots of the same color with a line (you could specify geom = "line" in a new line in the code chunk above to do this), you can see that the two virtual lines are not parallel (see below plot), suggesting the presence of an interaction. The difference in score between recognition and grammar tasks for Huntingtons patients (consider the vertical difference) is larger than the difference in score between recognition and grammar tasks for the Control patients. If those vertical differences were the same, there would be no interaction.
Provide key model results in a formatted table.
Use tab_model() from the sjPlot package.
Remember that you can rename your DV and IV labels by specifying dv.labels and pred.labels.
Interpret your results in the context of the research question and report your model in full.
Make reference to the interaction plot and regression table.
Full regression results including 95% Confidence Intervals are shown in Table 2. The \(F\)-test for model utility was significant \((F(5,24) = 13.62, p <.001)\), and the model explained approximately 68.5% of the variability in Scores.
The difference in scores between the recognition and grammar tasks, respectively measuring explicit and implicit memory, for amnesiac patients in comparison to controls was estimated to be -10 points, though this difference was not significantly different from zero \((t(24) = -0.85, p = .40)\).
The difference in scores between the recognition and grammar tasks, respectively measuring explicit and implicit memory, for Huntingtons patients in comparison to controls was significant, and indicated a difference of 40 points in explicit vs implicit memory performance \((t(24) = 3.41, p = .002)\). This interaction is visually presented in Figure 2.
Therefore, we have evidence to reject the null hypothesis (the difference in performance between explicit and implicit memory tasks does not significantly differ between patients with Huntingtons in comparison to Controls).