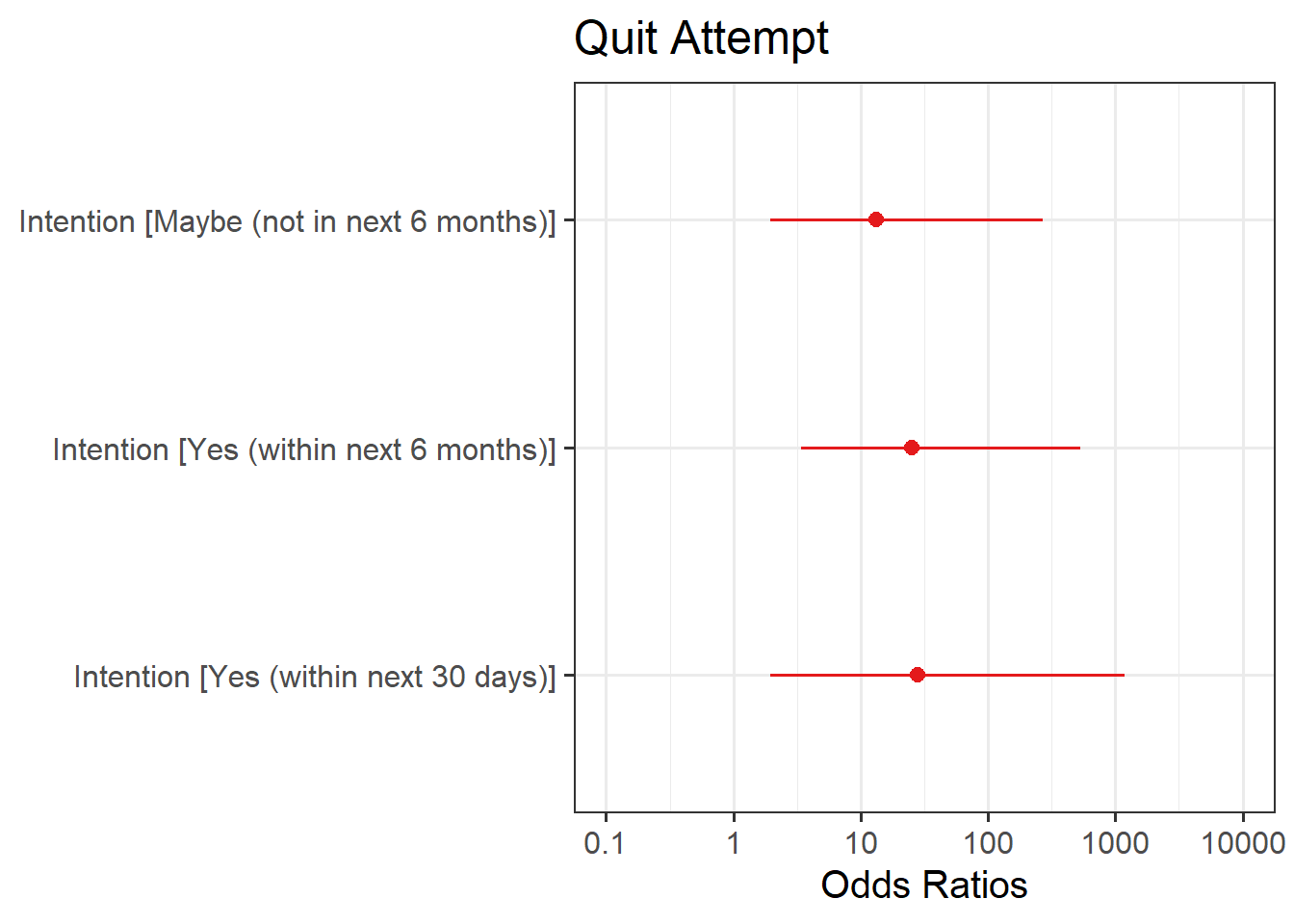| variable | description |
|---|---|
| S | Subject |
| Q | Quit Attempt, Wheter the subject made an attempt to quit using tobacco products (0 = no attempt to quit; 1 = attempted to quit). |
| I | Intention to Quit, with four levels: 1 = Never intend to quit; 2 = May intend to quit but not in the next 6 months; 3 = Intend to quit in the next 6 months; 4 = Intend to quit in the next 30 days |
More Logistic Regression
Learning Objectives
At the end of this lab, you will:
- Understand when to use a logistic model
- Understand how to fit and interpret a logistic model
- Understand how to evaluate model fit
What You Need
- Be up to date with lectures
- Have completed previous lab exercises from Week 6
Required R Packages
Remember to load all packages within a code chunk at the start of your RMarkdown file using library(). If you do not have a package and need to install, do so within the console using install.packages(" "). For further guidance on installing/updating packages, see Section C here.
For this lab, you will need to load the following package(s):
- tidyverse
- patchwork
- kableExtra
- psych
- sjPlot
Lab Data
You can download the data required for this lab here or read it in via this link https://uoepsy.github.io/data/QuitAttempts.csv.
Study Overview
Research Question
Is attempting to quit tobacco products associated with an individuals intentions?
Setup
- Create a new RMarkdown file
- Load the required package(s)
- Read in the QuitAttempts dataset into R, assigning it to an object named
smoke
Examine the dataset, and perform any necessary and appropriate data management steps.
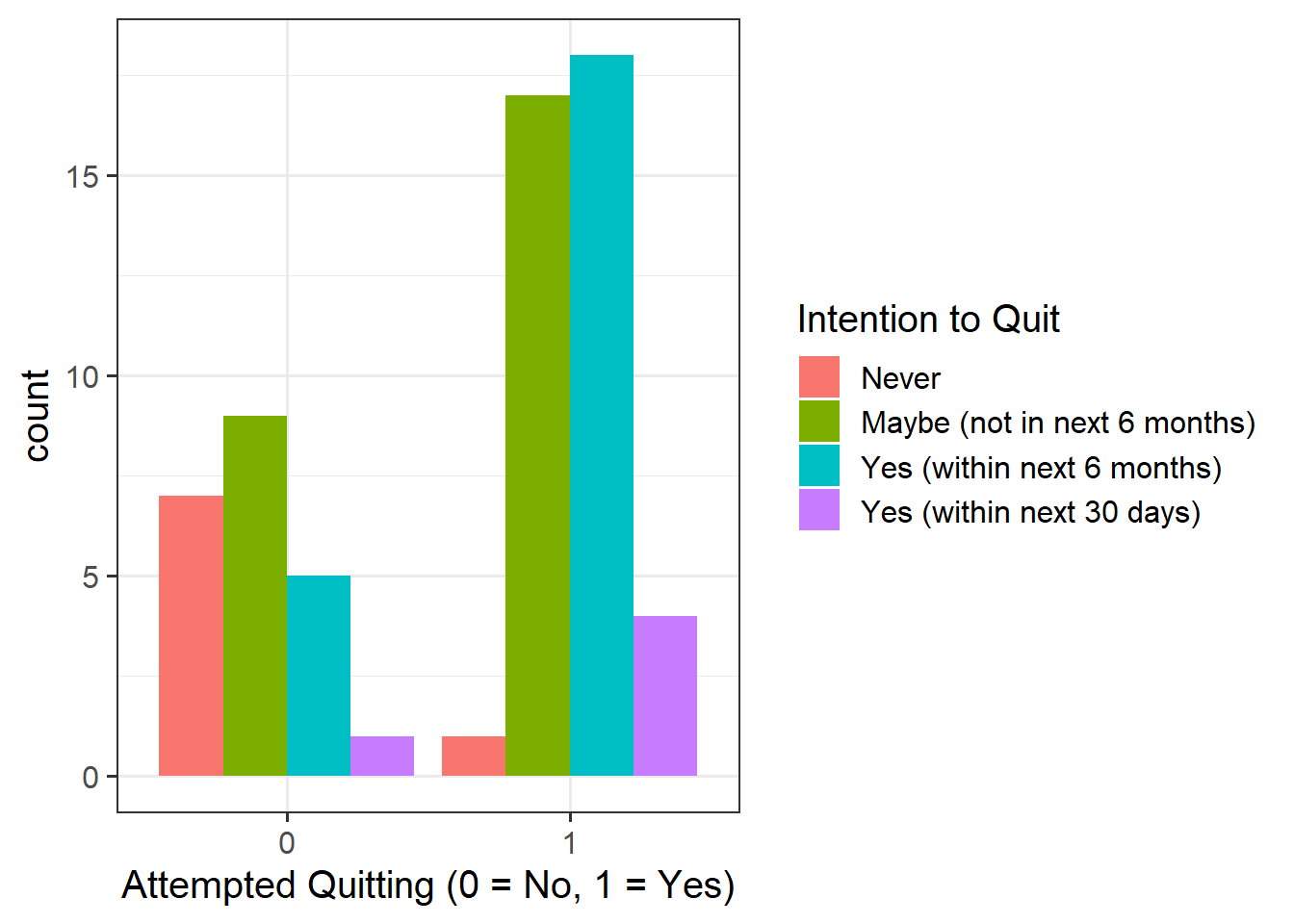
Provide a table of descriptive statistics and visualise your data.
Remember to interpret your plot in the context of the study.
- For your table of descriptive statistics, since we have two categorical variables, the
select()andtable()functions will come in handy here. - Recall that when visualising a continuous outcome across several groups,
geom_boxplot()may be most appropriate to use. - For your visualisations, you will need to specify
as_factor()when plotting theQuit_Attemptvariable since this is numeric, but we want it to be treated as a factor only for plotting purposes - Make sure to comment on any observed differences among the sample means of the four treatment conditions.
Fit your model using glm(), and assign it as an object with the name “smoke_mdl1”.
Interpret your coefficients in the context of the study. When doing so, it may be useful to translate the log-odds back into odds.
The opposite of the natural logarithm is the exponential (see here for more details if you are interested), and in R these functions are log() and exp().
Recall that we can obtain our parameter estimates using various functions such as summary(),coef(), coefficients(), etc. Thus, we want to exponentiate the coefficients from our model in order to translate them back from log-odds.
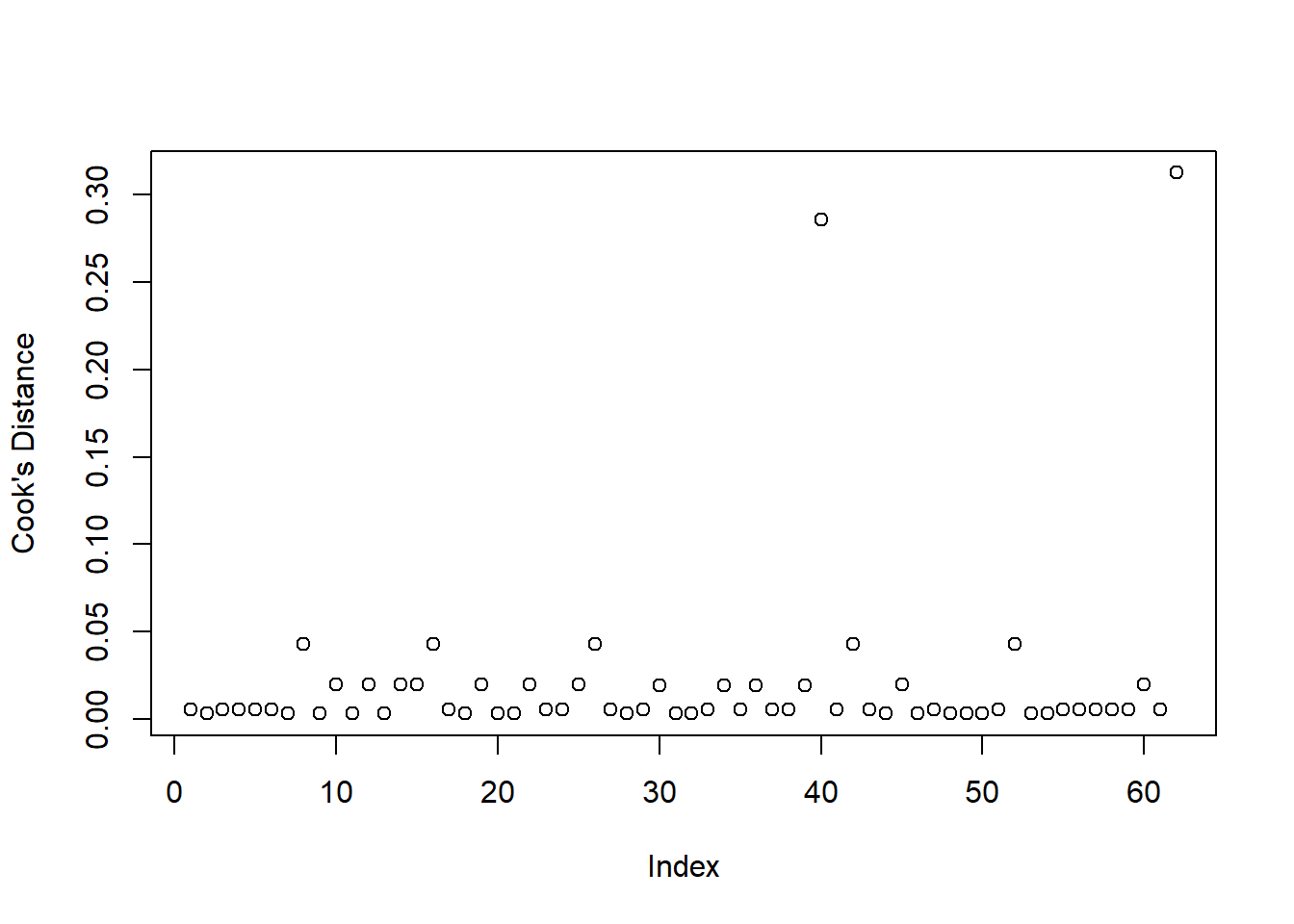
Examine the below plot to determine if the deviance residuals raise concerns about outliers:
Based on this plot, are there any residuals of concern? Are there any additional plots you could check to determine if there are influential observations?
Deviance Residuals
Because logistic regression models don’t have the same expected error distribution (we don’t expect residuals to be normally distributed around the mean, with constant variance), checking the assumptions of logistic regression is a little different.
Typically, we look at the “deviance residuals”. But we don’t examine plots for patterns, we simply examine them for potentially outlying observations. If we use a standardised residual, it makes it easier to explore extreme values as we expect most residuals to be within -2, 2 or -3, 3 (depending on how strict we feel).
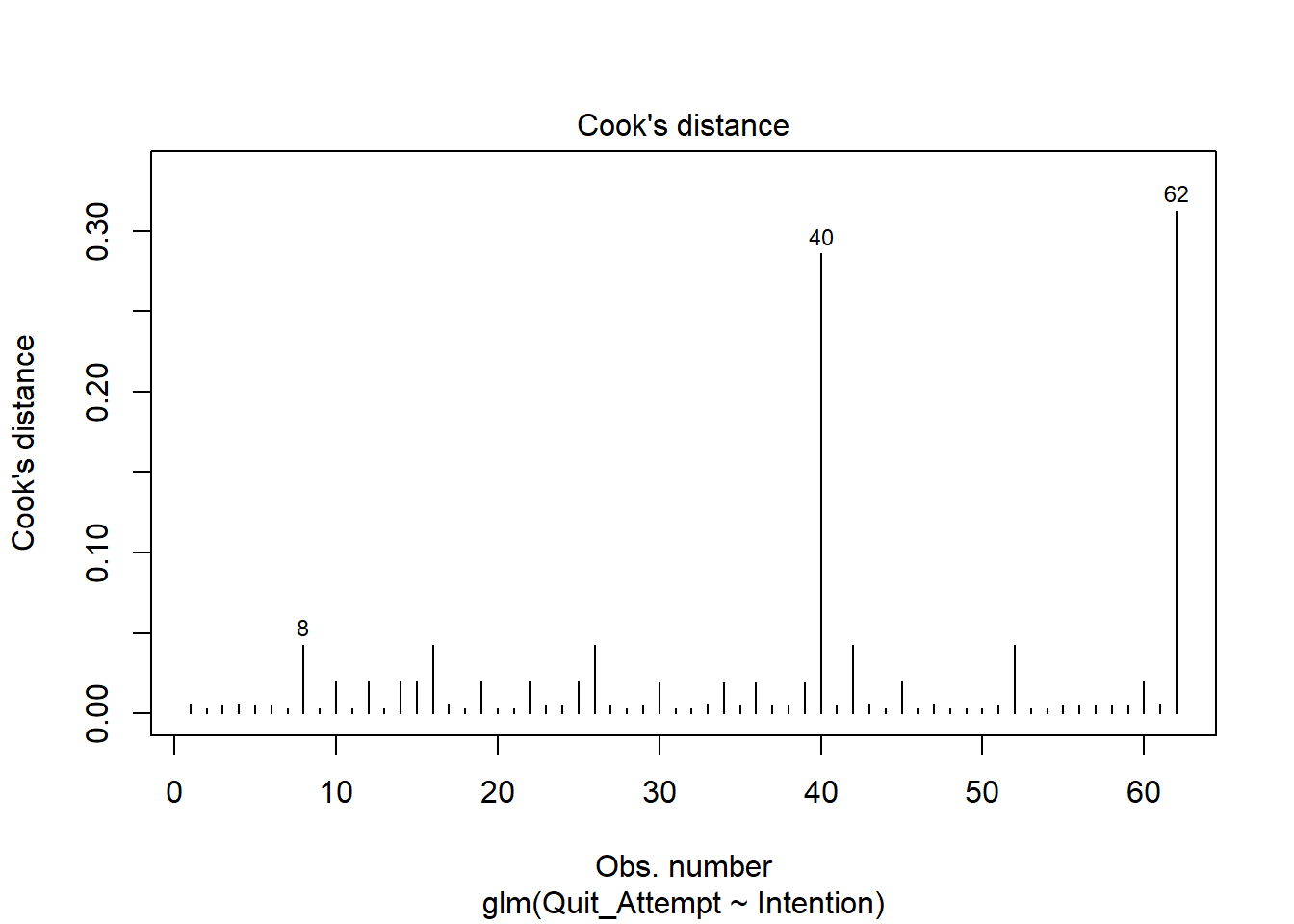
Cook’s Distance
To check for influential observations, we can use cooks.distance(), and plot this using plot(). Alternatively, you can specify which = 4 when plotting your fitted model. In logistic regression, we can use the arbitrary cut-offs of 0.5 (moderately influential) or 1 (highly influential) to describe influential points.
Perform a Deviance goodness-of-fit test to compare your fitted model to the null.
\[ \begin{aligned} M_0 &: \qquad \log \left( \frac{p}{1 - p}\right) = \beta_0 \\ M_1 &: \qquad \log \left( \frac{p}{1 - p}\right) = \beta_0 + \beta_1 Intention2 + \beta_2 Intention3 + \beta_3 Intention4 \end{aligned} \]
Report which model you think best fits the data.
Consider whether or not your models are nested. The flowchart in Question 10 of the Semester 2 Week 1 lab may be helpful to revisit.
Check the AIC and BIC values for smoke_mdl0 and smoke_mdl1 - which model should we prefer?
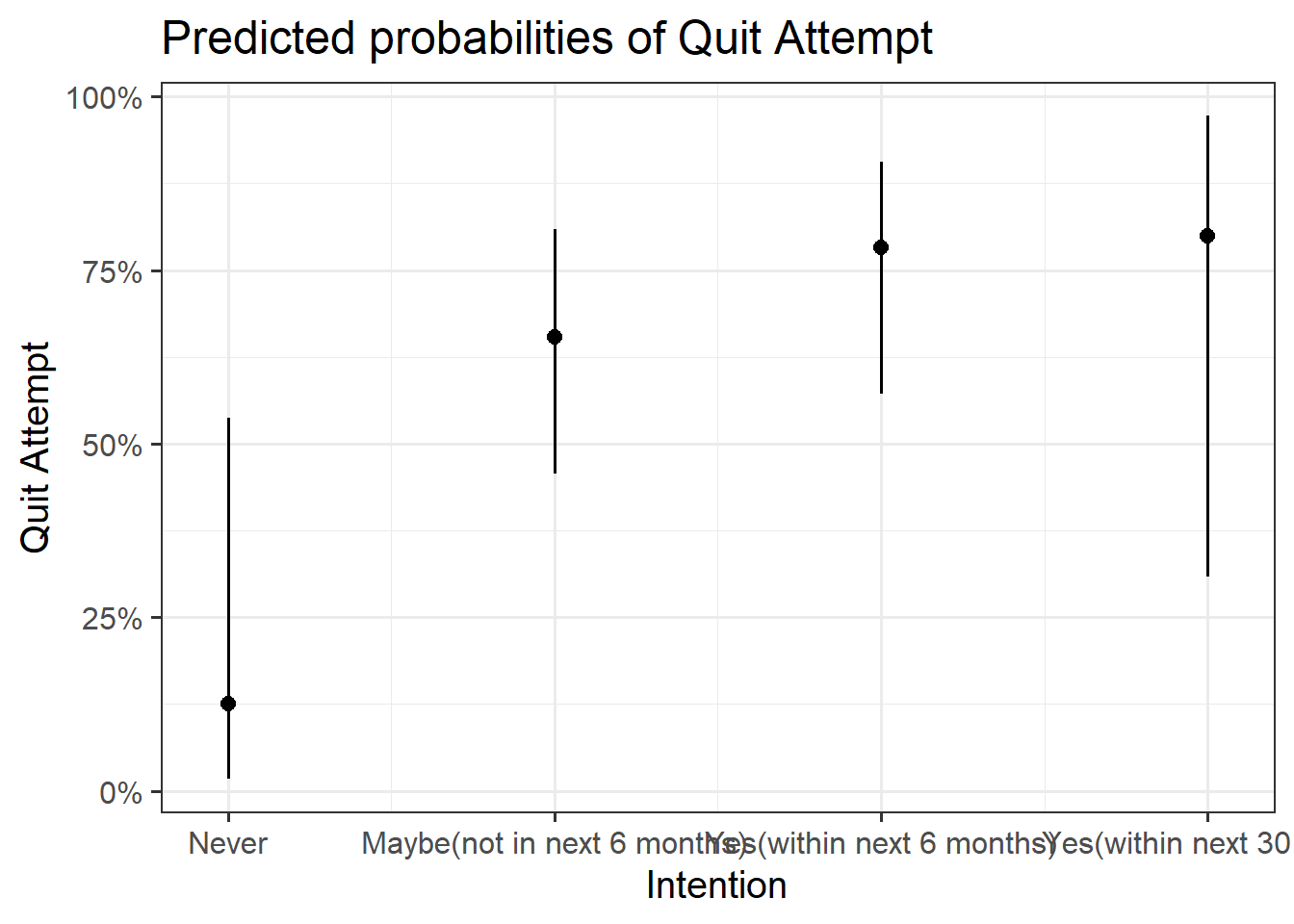
Plot the following:
- predicted model estimates
- predicted probability
Here you will need to use plot_model() from the sjPlot package like you did back in Semester 1. To get your estimates, you will need to specify type = "est", and for predicted probability, type = "eff".
Provide key model results in a formatted table.
Use tab_model() from the sjPlot package.
Remember that you can rename your DV and IV labels by specifying dv.labels and pred.labels.
Interpret your results in the context of the research question and report your model in full.
Footnotes
Kalkhoran, S., Grana, R. A., Neilands, T. B., & Ling, P. M. (2015). Dual use of smokeless tobacco or e-cigarettes with cigarettes and cessation. American Journal of Health Behavior, 39(2), 277–284. https://doi.org/10.5993/AJHB.39.2.14↩︎