Rows: 30 Columns: 3
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
dbl (3): Diagnosis, Task, Y
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Interactions II: Cat x Cat
Learning Objectives
At the end of this lab, you will:
- Be able to interpret a categorical \(\times\) categorical interaction.
- Visualize and probe interactions.
- Be able to read interaction plots.
What You Need
You will need to have completed [Lab X].
Required R Packages
Remember to load all packages within a code chunk at the start of your RMarkdown file using library(). If you do not have a package and need to install, do so within the console using install.packages(" "). For further guidance on installing/updating packages, see Section C here.
For this lab, you will need to load the following package(s):
- tidyverse
- sjPlot
- emmeans
Lab Data
You can download the data required for this lab here or read it in via this link https://uoepsy.github.io/data/cognitive_experiment_3_by_2.csv
Study Overview
Research Question
Specify specific question
A group of researchers wants to test an hypothesised theory according to which the difference in performance between explicit and implicit memory tasks will be greatest for Huntington patients in comparison to controls. On the other hand, the difference in performance between explicit and implicit memory tasks will not significantly differ between patients with amnesia in comparison to controls.
Setup
- Create a new RMarkdown file
- Load the required package(s)
- Read the cognitive_experiment_3_by_2 dataset into R, assigning it to an object named
cog.
Exercises
Examine the dataset, and perform all the appropriate data management steps:
- Convert categorical variables to factors
- Label appropriately factors to aid with your model interpretations
- If needed, provide better variable names
Choose appropriate reference levels for the factors.
Specify a multiple regression model to test the research hypothesis.
Fit the specified model using lm(), and store the model in an object named mdl_int.
Note: Fortunately, R computes the dummy variables for us! Thus. each row in the summary of the model will correspond to one of the estimated \(\beta\)’s in the equation above.
Create a table of group means, and map each coefficient to the group means.
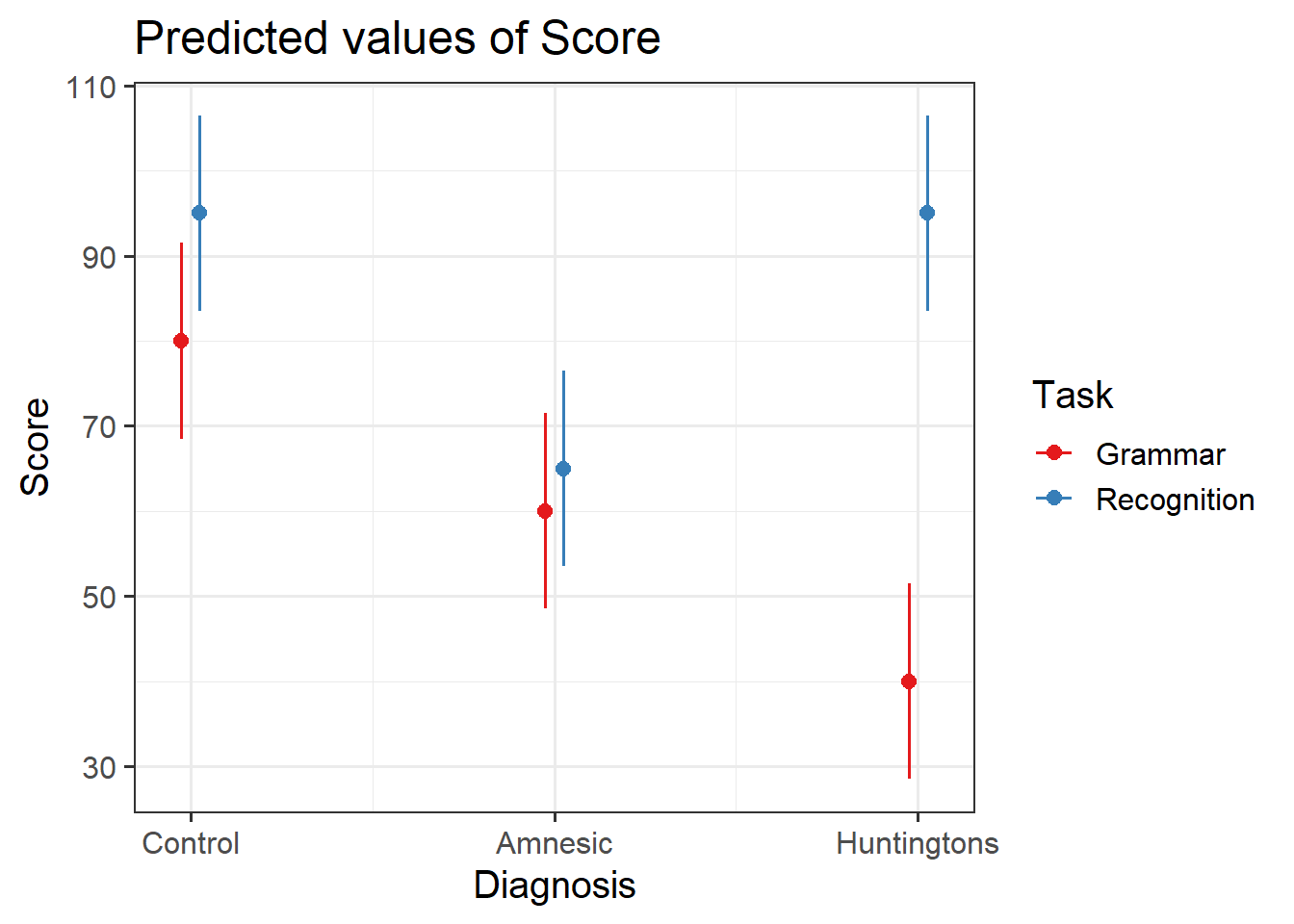
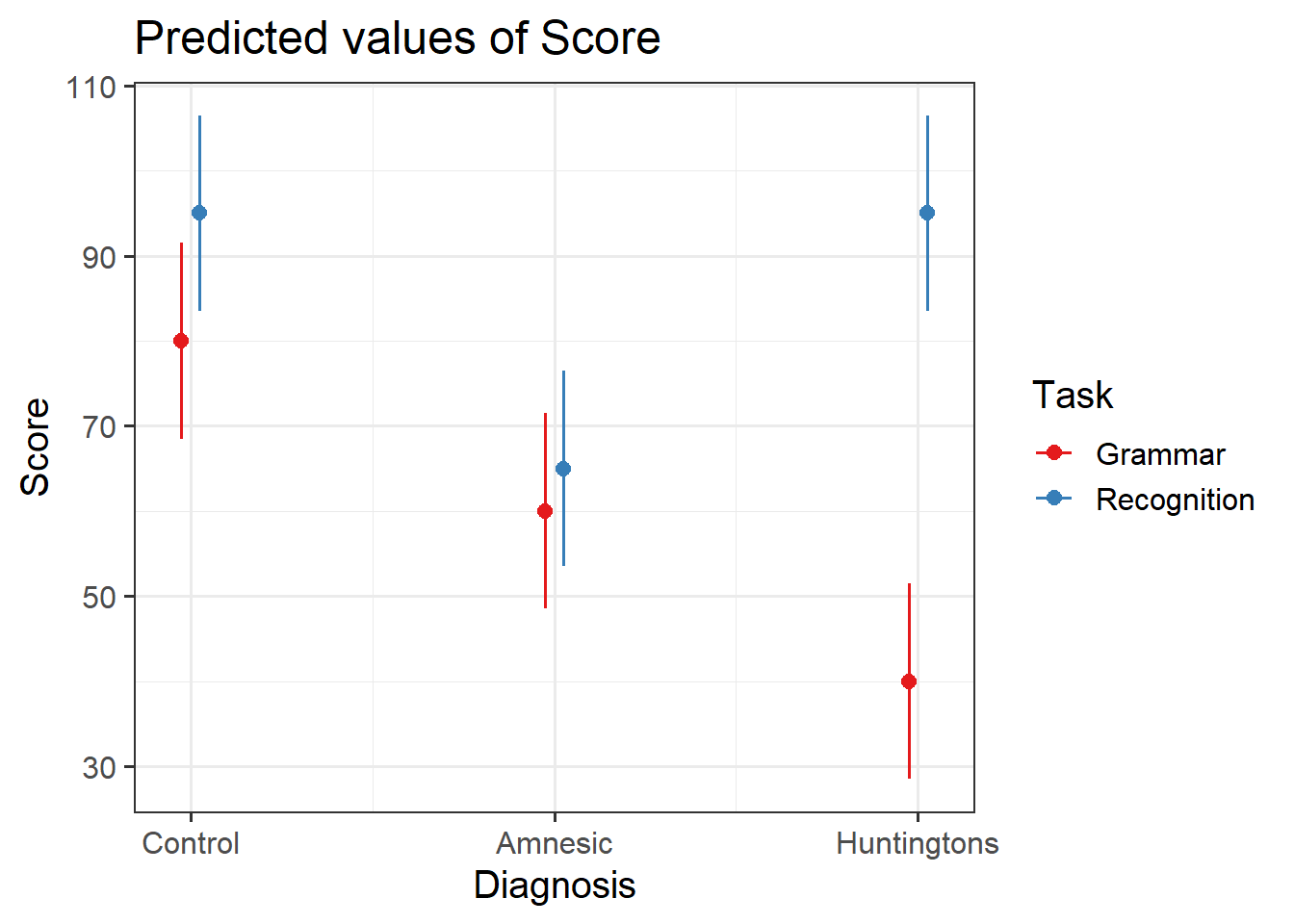
Use the function plot_model() (from the sjPlot package) to investigate the interactions.
Hint: Remember from last week that the default behaviour of plot_model() is to plot the parameter estimates and their confidence intervals. This is where type = " " will come handy.
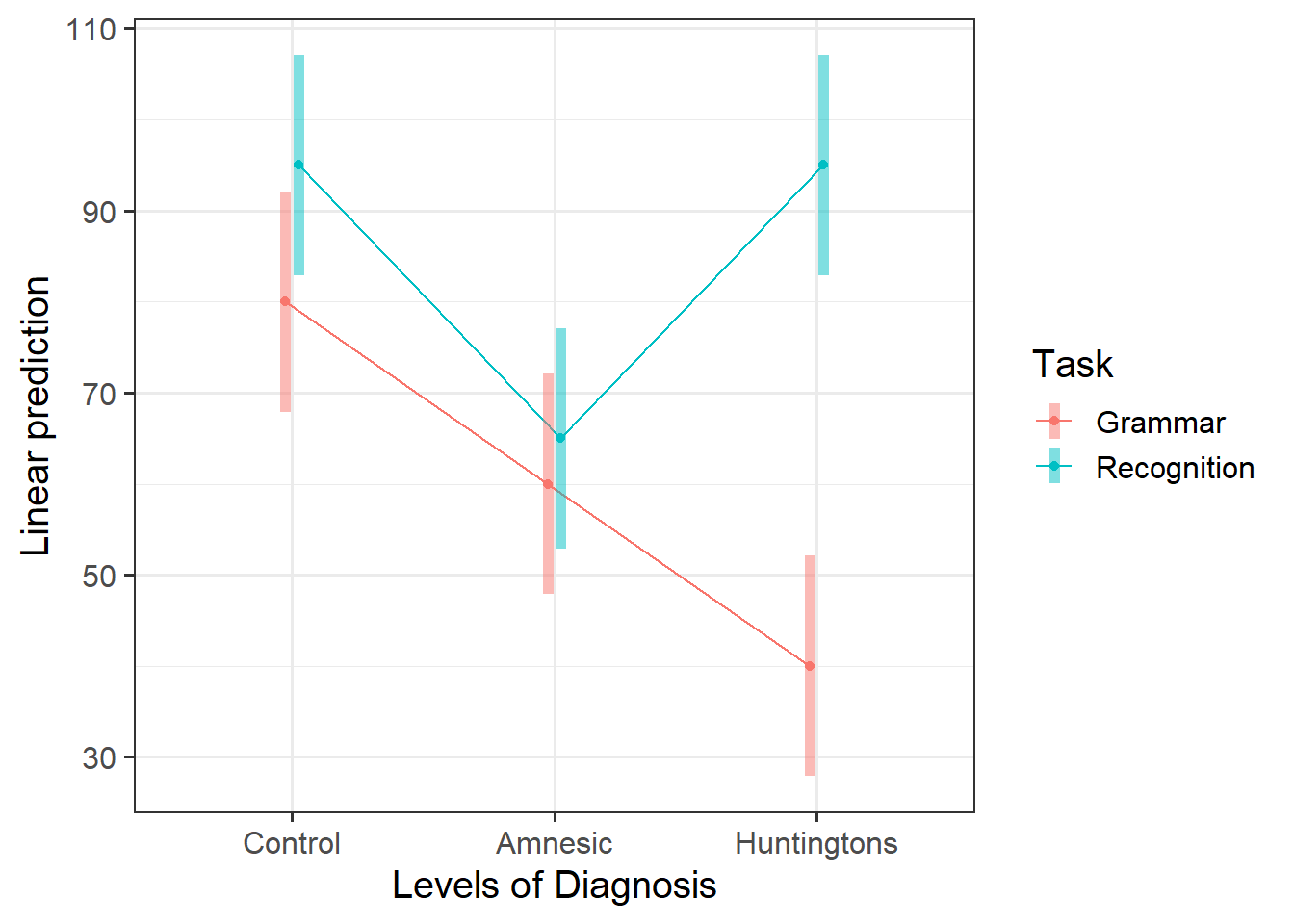
An alternative to plot_model() is the function emmip() (from the emmeans package) which stands for EMMeans Interaction Plot. Use the function emmip() to investigate the interactions.
Hint: You must provide as the first argument the fitted model, then a special formula saying what to plot on the x axis and what to use to differentiate the colors: colors ~ x, and then CIs = TRUE tells the function to display uncertainty intervals.
Visualising the interactions
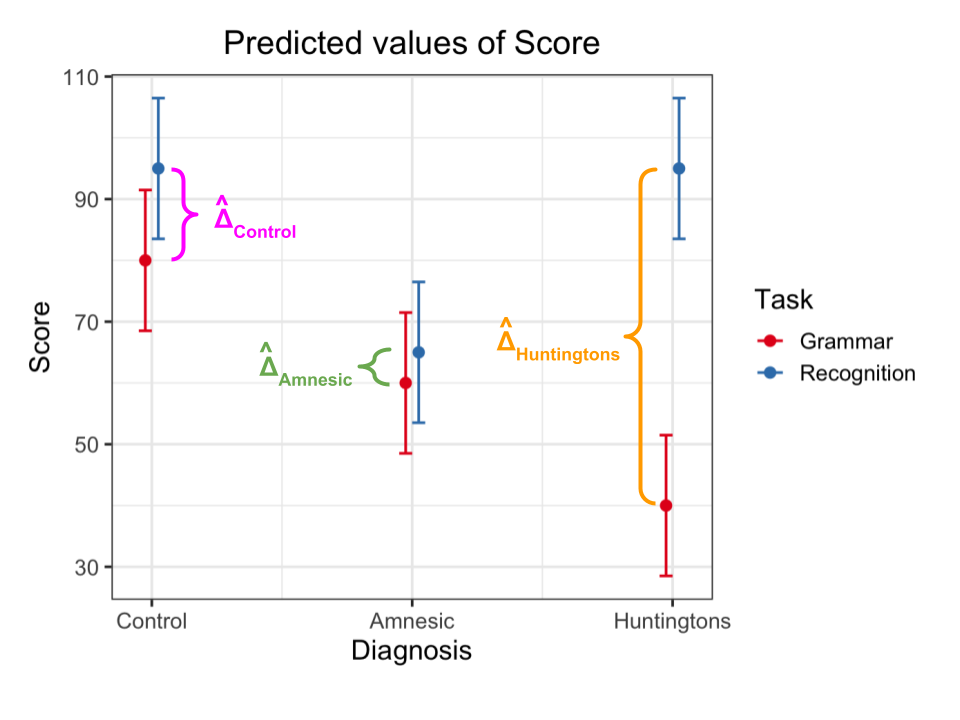
In the interaction plot above you can see three highlighted differences, where the differences are denoted with the Greek letter \(\Delta\) (“delta”) with a hat on top, \(\hat \Delta\), to denote that those are estimates for the unknown population differences based on the available sample data. The corresponding population differences are unknown as we don’t have the data for the entire population, and they are denoted with a \(\Delta\) without a hat on top.
You can see highlighted:
- The difference in the mean score between Recognition and Grammar for Control patients, \(\hat \Delta_{\text{Control}}\)
- The difference in the mean score between Recognition and Grammar for Amnesic patients, \(\hat \Delta_{\text{Amnesic}}\)
- The difference in the mean score between Recognition and Grammar for Huntingtons patients, \(\hat \Delta_{\text{Huntingtons}}\)
An interaction is present if the effect of Task (i.e. the difference in mean score between Recognition and Grammar tasks) substantially varies across the possible values for Diagnosis. That is, if the difference for Amnesic is not the same as that for Control, or if the difference for Huntingtons is not the same as that for Control, or both.
The model summary returns two rows for the interactions. Let’s focus on this row:
## DiagnosisAmnesic:TaskRecognition -10.000 11.719 -0.853 0.40192 It returns an estimate for the difference
\[ \hat \beta_4 = \hat \Delta_{\text{Amnesic}} - \hat \Delta_{\text{Control}} \]
and also performs a test for the hypothesis that the differences are equal in the population:
\[ H_0: \Delta_{\text{Amnesic}} = \Delta_{\text{Control}} \\ H_1: \Delta_{\text{Amnesic}} \neq \Delta_{\text{Control}} \]
or, equivalently, that:
\[ H_0: \Delta_{\text{Amnesic}} - \Delta_{\text{Control}} = 0 \\ H_1: \Delta_{\text{Amnesic}} - \Delta_{\text{Control}} \neq 0 \]
Let’s now focus on the last row:
## DiagnosisHuntingtons:TaskRecognition 40.000 11.719 3.413 0.00228 ** It returns an estimate for the difference
\[ \hat \beta_5 = \hat \Delta_{\text{Huntingtons}} - \hat \Delta_{\text{Control}} \]
and also performs a test for the hypothesis that the differences are equal in the population:
\[ H_0: \Delta_{\text{Huntingtons}} = \Delta_{\text{Control}} \\ H_1: \Delta_{\text{Huntingtons}} \neq \Delta_{\text{Control}} \]
or, equivalently, that:
\[ H_0: \Delta_{\text{Huntingtons}} - \Delta_{\text{Control}} = 0 \\ H_1: \Delta_{\text{Huntingtons}} - \Delta_{\text{Control}} \neq 0 \]
Interpret the model output in the context of the research hypothesis.
Footnotes
Some researchers may point out that a design where each person was assessed on both tasks might have been more efficient. However, the task factor in such design would then be within-subjects, meaning that the scores corresponding to the same person would be correlated. To analyse such design we will need a different method which is discussed next year!↩︎