Exploratory vs Confirmatory Data Analysis
LEARNING OBJECTIVES
- Understand the difference between exploratory vs confirmatory analyses
- Understand how to select models by comparing test-data MSE
- Understand how to compute MSE via k-fold cross validation
Wine Quality
This week’s lab explores wine quality based on physiochemical properties/attributes, using data that was collected between 2004 and 2007. Specifically, the data concern white and red vinho verde.
The Data
Download the following datasets about wine quality:
- Red wine:
https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv - White wine:
https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-white.csv
If you open them on your PC, you will notice that the data values are separated by semicolons, rather than colons. We have to tell R this by using read_delim to read a file with delimited values, and specify the delimiter by saying delim = ";":
library(tidyverse)
red <-
read_delim("https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv",
delim = ";")
white <-
read_delim("https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-white.csv",
delim = ";")We will also add a column to each dataset specifying the wine color:
red <- red %>% mutate(col = "red")
white <- white %>% mutate(col = "white")The bind_rows function is used to combine the datasets (as the name suggests, we will be combining by row). Let’s call the combined data wine:
wine <- bind_rows(
red %>% mutate(col="red"),
white %>% mutate(col="white")
)Inspect the data and check the dimensions. How many observations and how many variables are there?
Exploratory analysis
For this section of the lab, we do not have a fixed research question and/or hypothesis, but will instead be exploring associations among variables. We have randomly selected two variables (alcohol, and col) to predict the outcome quality:
| Variable | Description |
|---|---|
quality
|
Quality rating of wine - assessed on a scale ranging from 0 (very bad) to 10 (excellent). |
alcohol
|
Alcohol volume - measured as a percentage. |
col
|
Colour of wine - red or white. |
Subset the data to only include these three variables we want to explore further - quality, alcohol, col.
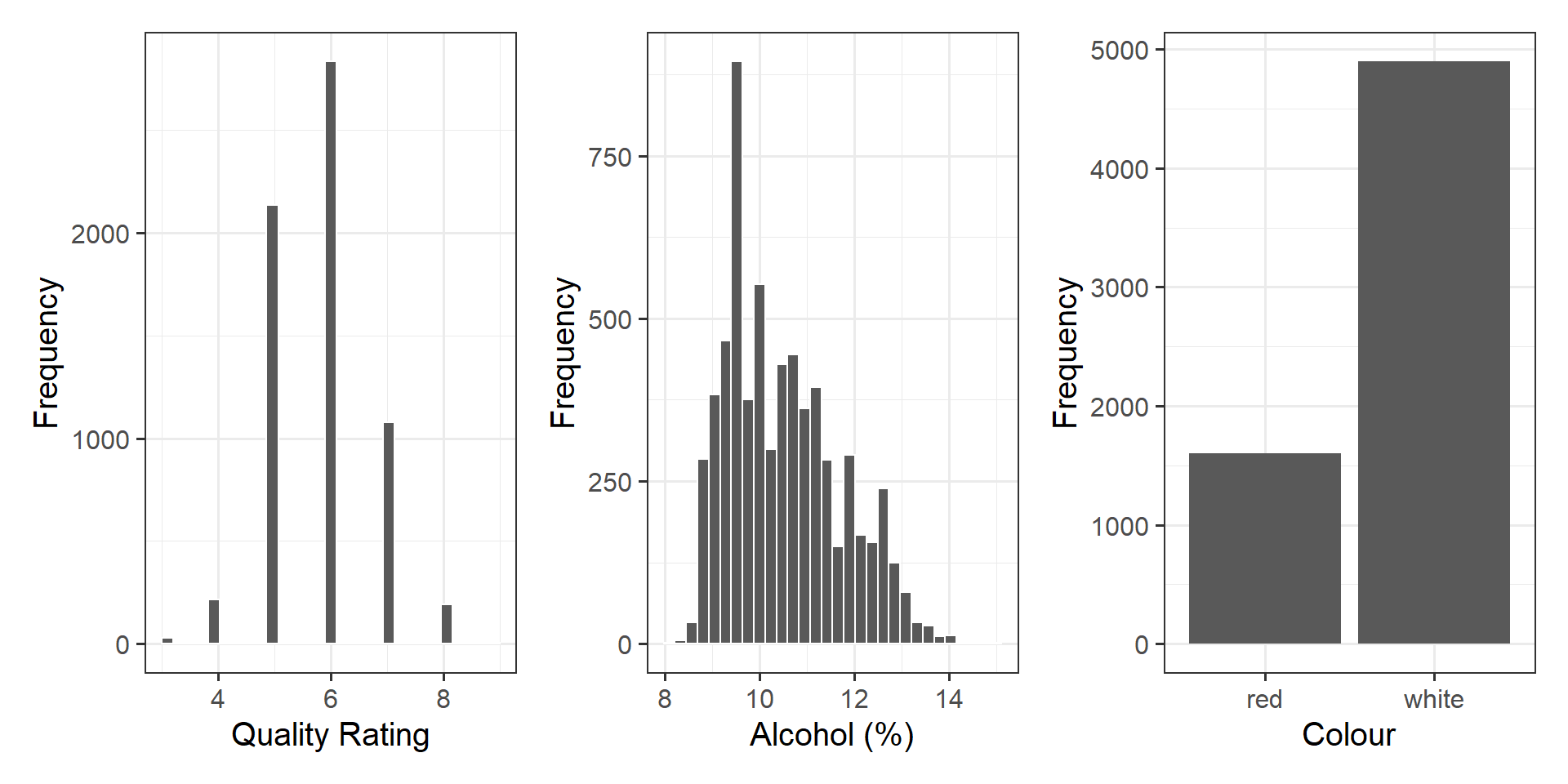
Visualise each variable individually, and note down your observations.
Produce a visualisation of the relationship between quality and alcohol. Consider either presenting with separate facets for wine colour, or add the colour argument within your aes() statement.
Our goal is to compare the following models to find if some of those variables are good predictors of quality ratings.
\[ \begin{aligned} M_A : \text{Quality} &= \beta_{A,0} + \beta_{A,1} \text{Alcohol} + \epsilon \\ M_B : \text{Quality} &= \beta_{B,0} + \beta_{B,1} \text{Col} + \epsilon \\ M_C : \text{Quality} &= \beta_{C,0} + \beta_{C,1} \text{Alcohol} + \beta_{C,2} \text{Col} + \epsilon \end{aligned} \]
To do so, we need to compute the k-fold cross validation MSE for each of those models:
\[ MSE_A = MSE(M_A) \\ MSE_B = MSE(M_B) \\ MSE_C = MSE(M_C) \]
We will begin by constructing the MSE for model A by hand, using \(k = 3\) folds.
As a first step, we need to randomise our data, and then divide into \(k\) groups, or “folds,” of roughly equal size. We will subset into 3 groups.
Recall that we have nrow(wine), and so need to create two datasets of 2166 observations, and one with 2165 observations.
We are going to work through the following models:
- \(MA_{1}\): trained on
bind_rows(wine2, wine3), tested onwine1 - \(MA_{2}\): trained on
bind_rows(wine1, wine3), tested onwine2 - \(MA_{3}\): trained on
bind_rows(wine1, wine2), tested onwine3
Recall model A involves fitting:
\[ M_A : \text{Quality} = \beta_{A,0} + \beta_{A,1} \text{Alcohol} + \epsilon \]
Calculate the test MSE on the observations in the fold that was held out. Remember that the MSE is the average squared distance between the observed and predicted values.
We now need to compute the same for the other 2 models too, in order to compare them.
For the next two models, we will use the cross-validation functions from the modelr package, as shown in the lecture.
In R, as demonstrated in the lectures, you can use the crossv_kfold() function from the modelr package to conduct K-Folding. You also need to use the map() function too.
library(modelr)
# example of three folds
CV <- crossv_kfold(wine, k = 3)
CV## # A tibble: 3 x 3
## train test .id
## <named list> <named list> <chr>
## 1 <resample [4,331 x 3]> <resample [2,166 x 3]> 1
## 2 <resample [4,331 x 3]> <resample [2,166 x 3]> 2
## 3 <resample [4,332 x 3]> <resample [2,165 x 3]> 3Important! We selected two variables at random from the original wine dataset. If you would like more practice, feel free to ‘explore’ the associations of other variables that you think sound intriguing!
mB <- map(CV$train, ~lm(quality ~ col, data = .))
mB## $`1`
##
## Call:
## lm(formula = quality ~ col, data = .)
##
## Coefficients:
## (Intercept) colwhite
## 5.6267 0.2642
##
##
## $`2`
##
## Call:
## lm(formula = quality ~ col, data = .)
##
## Coefficients:
## (Intercept) colwhite
## 5.6645 0.2033
##
##
## $`3`
##
## Call:
## lm(formula = quality ~ col, data = .)
##
## Coefficients:
## (Intercept) colwhite
## 5.617 0.258# helper function from lecture
get_pred <- function(model, test_data){
data = as.data.frame(test_data)
pred = add_predictions(data, model)
return(pred)
}
predB <- map2_df(mB, CV$test, get_pred, .id = "Run")
predB## # A tibble: 6,497 x 5
## Run quality alcohol col pred
## <chr> <dbl> <dbl> <chr> <dbl>
## 1 1 5 9.8 red 5.63
## 2 1 5 9.4 red 5.63
## 3 1 5 9.1 red 5.63
## 4 1 5 9.2 red 5.63
## 5 1 5 9.3 red 5.63
## 6 1 4 9 red 5.63
## 7 1 5 9.5 red 5.63
## 8 1 5 9.4 red 5.63
## 9 1 6 9.7 red 5.63
## 10 1 5 9.4 red 5.63
## # ... with 6,487 more rowsmse_B_folds <- predB %>%
group_by(Run) %>%
summarise(MSE = mean((quality - pred)^2))
mse_B_folds## # A tibble: 3 x 2
## Run MSE
## <chr> <dbl>
## 1 1 0.768
## 2 2 0.793
## 3 3 0.696mse_B <- mean(mse_B_folds$MSE)
mse_B## [1] 0.752463As you see, model B leads to a MSE of 0.75.
Calculate the MSE for model C using 3-fold cross validation.
Which model is the best model?
Confirmatory
You have already conducted many confirmatory analyses during the DAPR2 course - you have been asked to complete specific analyses, test pre-determined hypotheses, etc.
If you have a strong research question, and you approach the problem using an exploratory analysis approach, you may end up finding that the model with the lowest MSE is not a model that includes the variables that are required to test your hypothesis.
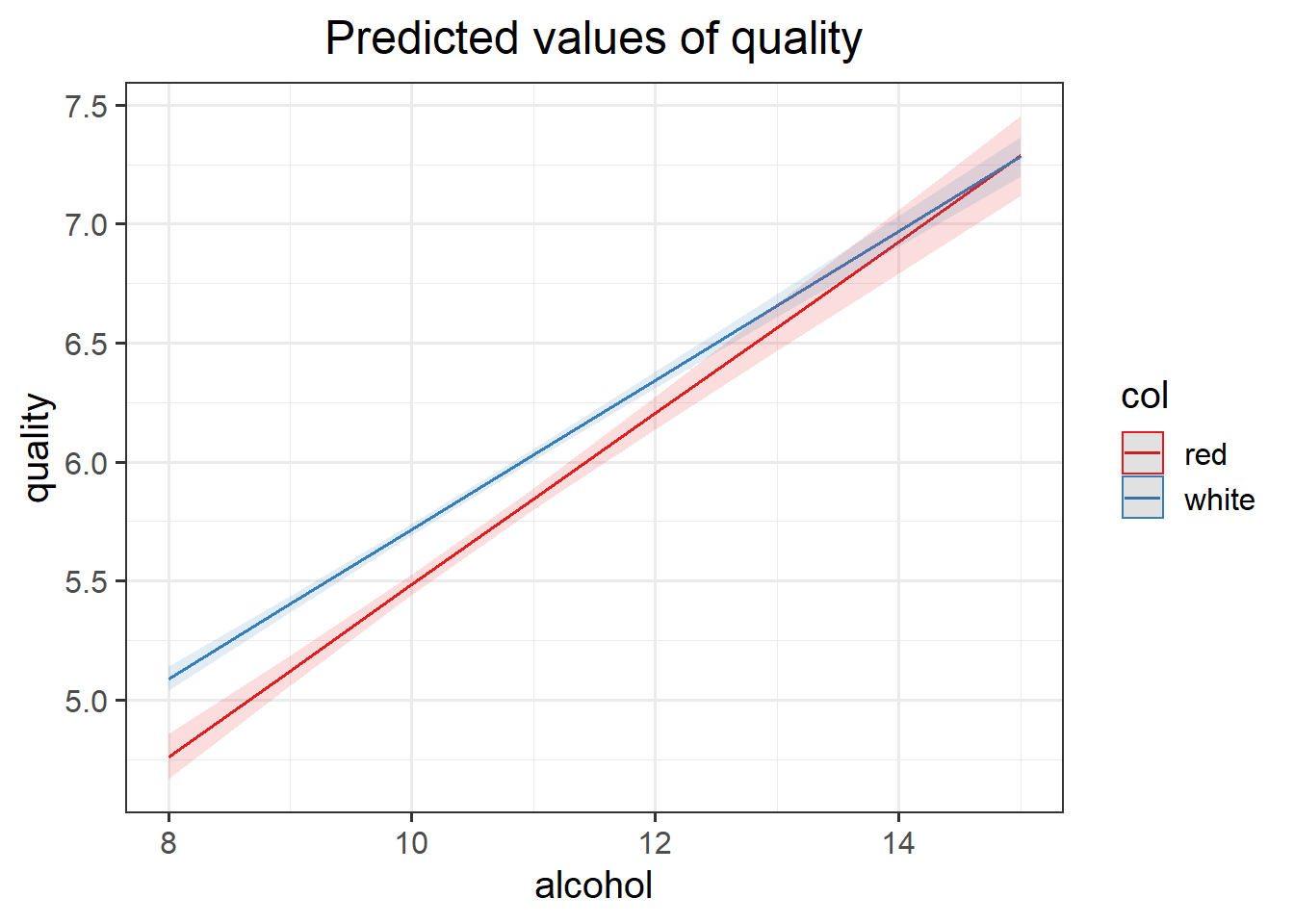
For example, suppose in the exploratory analysis above you also included a \(M_D\) that included the interaction between alcohol content and wine color. It could happen that the model with the lowest MSE was not \(M_D\), meaning that you would not be working with that model.
However, suppose you wanted to perform a confirmatory analysis aimed at testing whether the effect of alcohol percentage on wine quality rating is dependent on the color of the wine.
To answer such question your model must have the interaction term as the question directly relates to the interaction term.
Fit a model that can answer the stated research hypothesis, and interpret the results.