Assumptions, Multiple Comparisons, Corrections, & Write-Up Example
LEARNING OBJECTIVES
- Assess if a fitted model satisfies the assumptions of your model.
- Understand how to apply corrections available for multiple comparisons.
- Understand how to write-up and provide interpretation of a 2x2 factorial ANOVA.
Assumption Checks
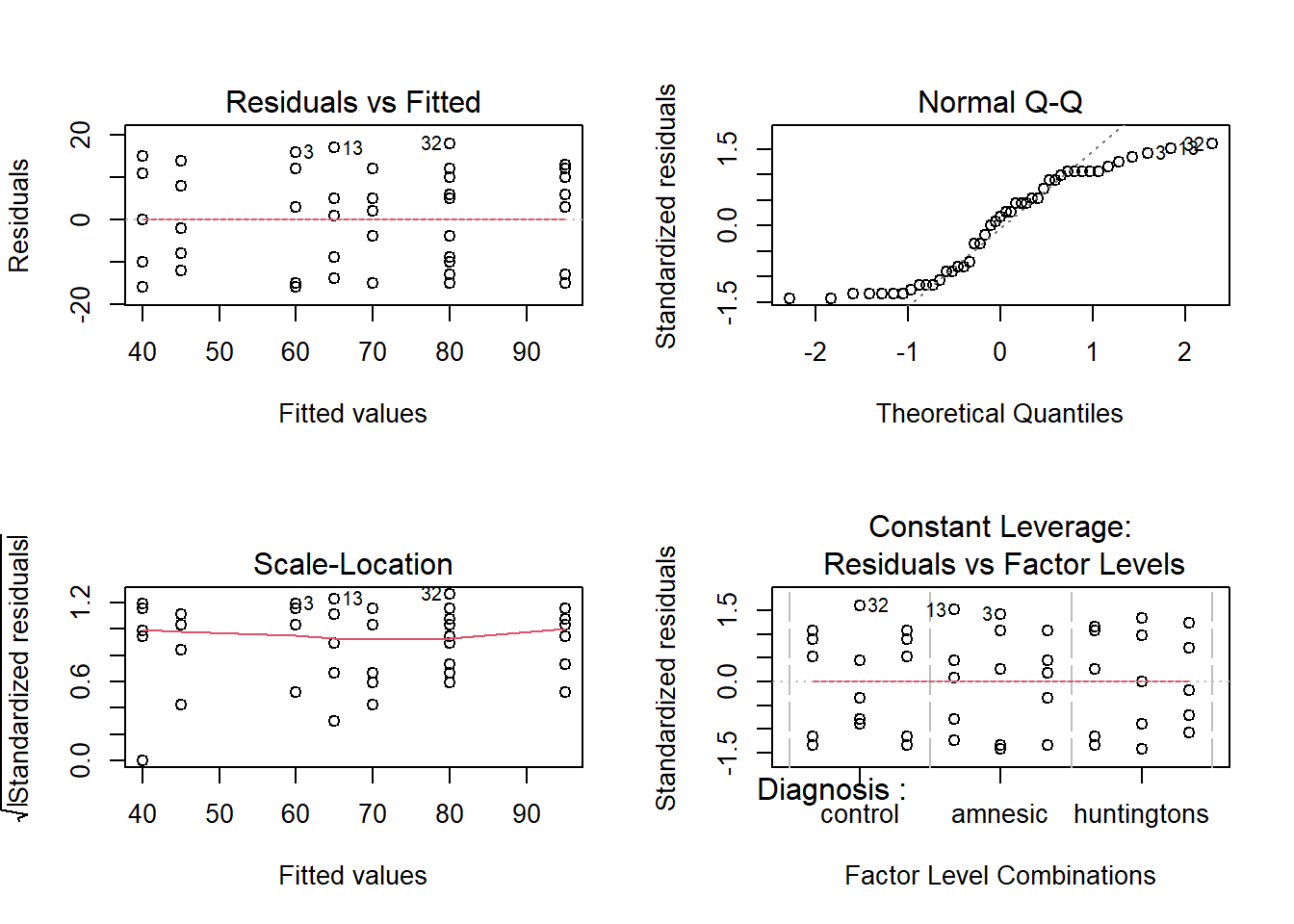
With our cog data model, we have yet to check our assumptions (bad practice for us leaving it so late!). The good news is that most are the same as for linear regression. The only difference is that linearity is now replaced by checking whether the errors in each group have mean zero.
Let’s check that the interaction model doesn’t violate the assumptions.
- Are the errors independent?
- Do the errors have a mean of zero?
- Are the group variances equal?
- Is the distribution of the errors normal?
WARNING
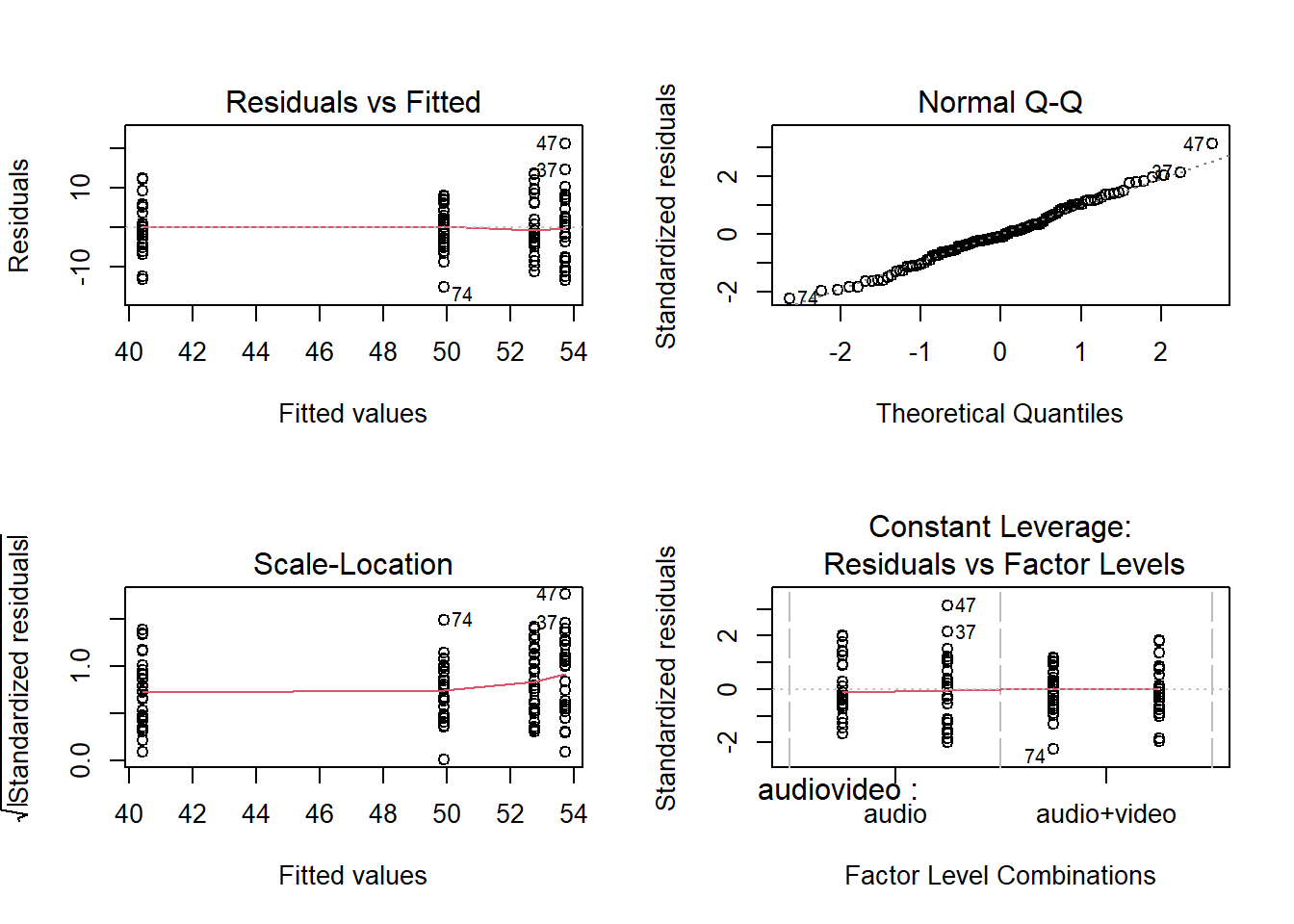
The residuals don’t look like a sample from a normal population. For this reason, we can’t trust the model results and we should not generalise the results to the population as the hypothesis tests to be valid require all the assumptions to be met, including normality.
We will nevertheless carry on and finish this example so that we can exploring the remaining functions relevant for carrying out multiple comparisons with adjustments with a two-way ANOVA.
Multiple Comparisons
In last week’s exercises we began to look at how we compare different groups, by using contrast analysis to conduct tests of specific comparisons between groups. We also saw how we might conduct pairwise comparisons, where we test all possible pairs of group means within a given set.
For instance, we can compare the means of the different diagnosis groups for each task:
emm_task <- emmeans(mdl_int, ~ Diagnosis | Task)
contr_task <- contrast(emm_task, method = 'pairwise')
contr_task## Task = recognition:
## contrast estimate SE df t.ratio p.value
## control - amnesic 30 7.94 36 3.776 0.0016
## control - huntingtons 0 7.94 36 0.000 1.0000
## amnesic - huntingtons -30 7.94 36 -3.776 0.0016
##
## Task = grammar:
## contrast estimate SE df t.ratio p.value
## control - amnesic 20 7.94 36 2.518 0.0424
## control - huntingtons 40 7.94 36 5.035 <.0001
## amnesic - huntingtons 20 7.94 36 2.518 0.0424
##
## Task = classification:
## contrast estimate SE df t.ratio p.value
## control - amnesic 10 7.94 36 1.259 0.4273
## control - huntingtons 35 7.94 36 4.406 0.0003
## amnesic - huntingtons 25 7.94 36 3.147 0.0091
##
## P value adjustment: tukey method for comparing a family of 3 estimatesor we can test all different combinations of task and diagnosis group (if that was something we were theoretically interested in, which is unlikely!) which would equate to conducting 36 comparisons!
emm_task <- emmeans(mdl_int, ~ Diagnosis * Task)
contr_task <- contrast(emm_task, method = 'pairwise')
contr_task## contrast estimate SE df t.ratio
## control recognition - amnesic recognition 30 7.94 36 3.776
## control recognition - huntingtons recognition 0 7.94 36 0.000
## control recognition - control grammar 15 7.94 36 1.888
## control recognition - amnesic grammar 35 7.94 36 4.406
## control recognition - huntingtons grammar 55 7.94 36 6.923
## control recognition - control classification 15 7.94 36 1.888
## control recognition - amnesic classification 25 7.94 36 3.147
## control recognition - huntingtons classification 50 7.94 36 6.294
## amnesic recognition - huntingtons recognition -30 7.94 36 -3.776
## amnesic recognition - control grammar -15 7.94 36 -1.888
## amnesic recognition - amnesic grammar 5 7.94 36 0.629
## amnesic recognition - huntingtons grammar 25 7.94 36 3.147
## amnesic recognition - control classification -15 7.94 36 -1.888
## amnesic recognition - amnesic classification -5 7.94 36 -0.629
## amnesic recognition - huntingtons classification 20 7.94 36 2.518
## huntingtons recognition - control grammar 15 7.94 36 1.888
## huntingtons recognition - amnesic grammar 35 7.94 36 4.406
## huntingtons recognition - huntingtons grammar 55 7.94 36 6.923
## huntingtons recognition - control classification 15 7.94 36 1.888
## huntingtons recognition - amnesic classification 25 7.94 36 3.147
## huntingtons recognition - huntingtons classification 50 7.94 36 6.294
## control grammar - amnesic grammar 20 7.94 36 2.518
## control grammar - huntingtons grammar 40 7.94 36 5.035
## control grammar - control classification 0 7.94 36 0.000
## control grammar - amnesic classification 10 7.94 36 1.259
## control grammar - huntingtons classification 35 7.94 36 4.406
## amnesic grammar - huntingtons grammar 20 7.94 36 2.518
## amnesic grammar - control classification -20 7.94 36 -2.518
## amnesic grammar - amnesic classification -10 7.94 36 -1.259
## amnesic grammar - huntingtons classification 15 7.94 36 1.888
## huntingtons grammar - control classification -40 7.94 36 -5.035
## huntingtons grammar - amnesic classification -30 7.94 36 -3.776
## huntingtons grammar - huntingtons classification -5 7.94 36 -0.629
## control classification - amnesic classification 10 7.94 36 1.259
## control classification - huntingtons classification 35 7.94 36 4.406
## amnesic classification - huntingtons classification 25 7.94 36 3.147
## p.value
## 0.0149
## 1.0000
## 0.6257
## 0.0026
## <.0001
## 0.6257
## 0.0711
## <.0001
## 0.0149
## 0.6257
## 0.9993
## 0.0711
## 0.6257
## 0.9993
## 0.2575
## 0.6257
## 0.0026
## <.0001
## 0.6257
## 0.0711
## <.0001
## 0.2575
## 0.0004
## 1.0000
## 0.9367
## 0.0026
## 0.2575
## 0.2575
## 0.9367
## 0.6257
## 0.0004
## 0.0149
## 0.9993
## 0.9367
## 0.0026
## 0.0711
##
## P value adjustment: tukey method for comparing a family of 9 estimates
Why does the number of tests matter?
But this error-rate applies to each statistical hypothesis we test. So if we conduct an experiment in which we plan on conducting lots of tests of different comparisons, the chance of an error being made increases substantially. Across the family of tests performed that chance will be much higher than 5%.1
Each test conducted at \(\alpha = 0.05\) has a 0.05 (or 5%) probability of Type I error (wrongly rejecting the null hypothesis). If we do 9 tests, that experimentwise error rate is \(\alpha_{ew} \leq 9 \times 0.05\), where 9 is the number of comparisons made as part of the experiment.
Thus, if nine independent comparisons were made at the \(\alpha = 0.05\) level, the experimentwise Type I error rate \(\alpha_{ew}\) would be at most \(9 \times 0.05 = 0.45\). That is, we could wrongly reject the null hypothesis on average 45 times out of 100. To make this more confusing, many of the tests in a family are not independent (see the lecture slides for the calculation of error rate for dependent tests).
Here, we go through some of the different options available to us to control, or ‘correct’ for this problem.
Corrections
Bonferroni
Load the data from last week, and re-acquaint yourself with it.
Provide a plot of the Diagnosis*Task group mean scores.
The data is at https://uoepsy.github.io/data/cognitive_experiment.csv.
Fit the interaction model, using lm().
Pass your model to the anova() function, to remind yourself that there is a significant interaction present.
There are various ways to make nice tables in RMarkdown.
Some of the most well known are:
- The knitr package has
kable()
- The pander package has
pander()
Pick one (or find go googling and find a package you like the look of), install the package (if you don’t already have it), then try to create a nice pretty ANOVA table rather than the one given by anova(model).
As in the previous week’s exercises, let us suppose that we are specifically interested in comparisons of the mean score across the different diagnosis groups for a given task.
Edit the code below to obtain the pairwise comparisons of diagnosis groups for each task. Use the Bonferroni method to adjust for multiple comparisons, and then obtain confidence intervals.
library(emmeans)
emm_task <- emmeans(mdl_int, ? )
contr_task <- contrast(emm_task, method = ?, adjust = ? )
adjusting \(\alpha\), adjusting p
In the lecture we talked about adjusting the \(\alpha\) level (i.e., instead of determining significance at \(p < .05\), we might adjust and determine a result to be statistically significant if \(p < .005\), depending on how many tests are in our family of tests).
Note what the functions in R do is adjust the \(p\)-value, rather than the \(\alpha\). The Bonferroni method simply multiplies the ‘raw’ p-value by the number of the tests.
In question 4 above, there are 9 tests being performed, but there are 3 in each ‘family’ (each Task).
Try changing your answer to question 4 to use adjust = "none", rather than "bonferroni", and confirm that the p-values are 1/3 of the size.
Šídák
The Sidak approach is slightly less conservative than the Bonferroni adjustment. Doing this with the emmeans package is easy, can you figure out how?
Hint: you just have to change the adjust argument in contrast() function.
Tukey
Like with Šídák, in R we can easily change to Tukey. Conduct pairwise comparisons of the scores of different Diagnosis groups on different Task types (i.e., the interaction), and use the Tukey adjustment.
Scheffe
Run the same pairwise comparison as above, but this time with the Scheffe adjustment.
When to use Which Correction
Bonferroni
- Use Bonferroni’s method when you are interested in a small number of planned contrasts (or pairwise comparisons).
- Bonferroni’s method is to divide alpha by the number of tests/confidence intervals.
- Assumes that all comparisons are independent of one another.
- It sacrifices slightly more power than Tukey’s method (discussed below), but it can be applied to any set of contrasts or linear combinations (i.e., it is useful in more situations than Tukey).
- It is usually better than Tukey if we want to do a small number of planned comparisons.
Šídák
- (A bit) more powerful than the Bonferroni method.
- Assumes that all comparisons are independent of one another.
- Less common than Bonferroni method, largely because it is more difficult to calculate (not a problem now we have computers).
Tukey
- It specifies an exact family significance level for comparing all pairs of treatment means.
- Use Tukey’s method when you are interested in all (or most) pairwise comparisons of means.
Scheffe
- It is the most conservative (least powerful) of all tests.
- It controls the family alpha level for testing all possible contrasts.
- It should be used if you have not planned contrasts in advance.
- For testing pairs of treatment means it is too conservative (you should use Bonferroni or Šídák).
In R, you can easily change which correction you are using via the adjust = argument.
Lie Detection Experiment - Write Up Example
In this section of the lab, you will be presented with a research question, and tasked with writing up and presenting your analyses. Try to write three complete sections - Analysis Strategy, Results, and Discussion. Make sure to familiarise yourself with the data available in the codebook below. We will use the questions to go through the analysis step by step, before writing up. Please note that the lab includes short example write ups that may not be complete for every question you are asked. It is intended to give you a sense of the style. Think about the steps you need to complete in order to answer the research question, and the order in which you should complete these. Under the solutions are the code chunks used to complete the steps outlined, and the write up section examples follow at the end.
Research Question
Do Police training materials and the mode of communication influence the accuracy of veracity judgements?
Step 1 is always to read in the data, then to explore, check, describe, and visualise it.
- Load the data
- Examine the data
- Check coding of variables (ie., are categorical variables factors?)
- Produce a descriptives table for the variables of interest
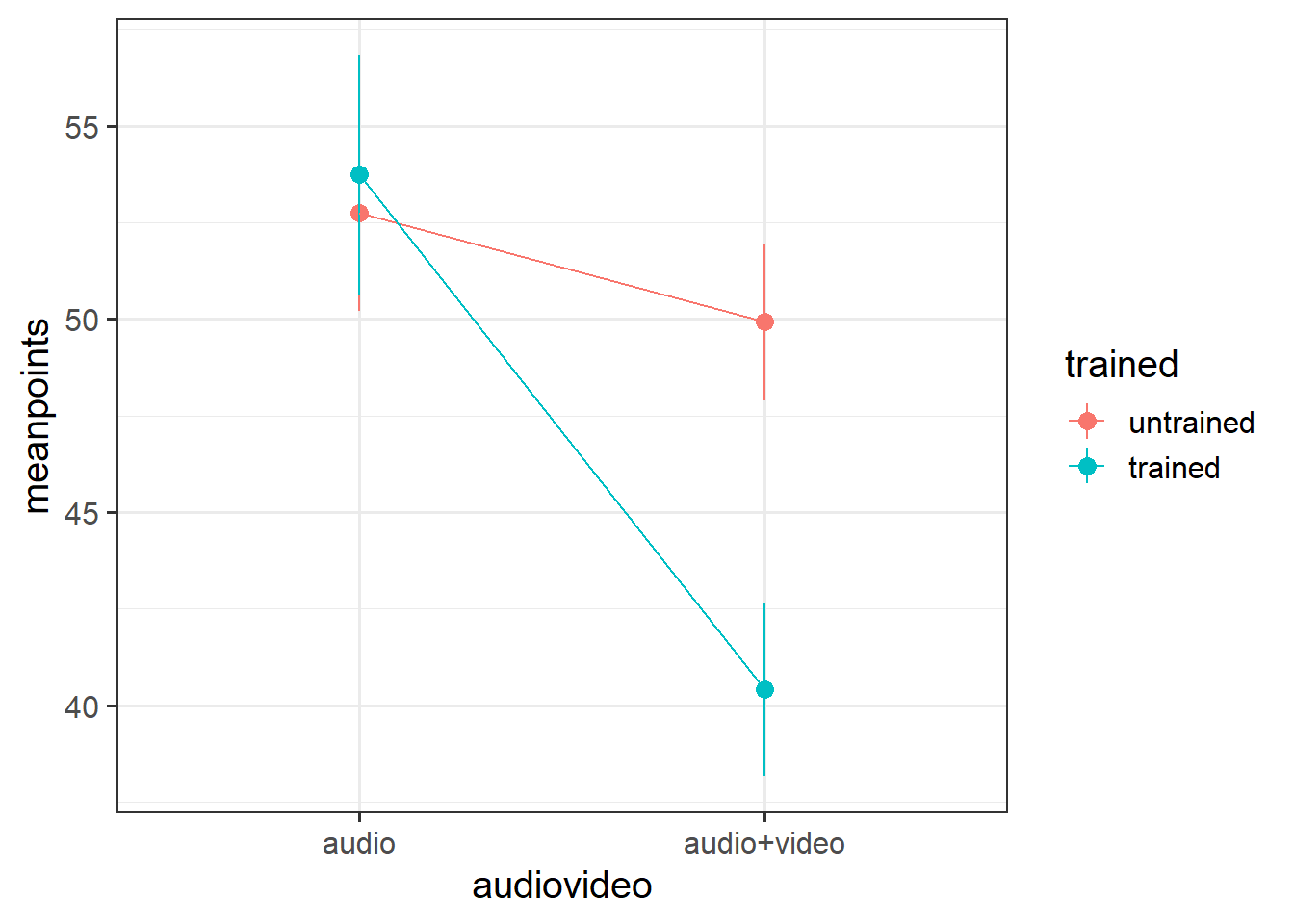
- Produce a plot showing the mean points for each condition
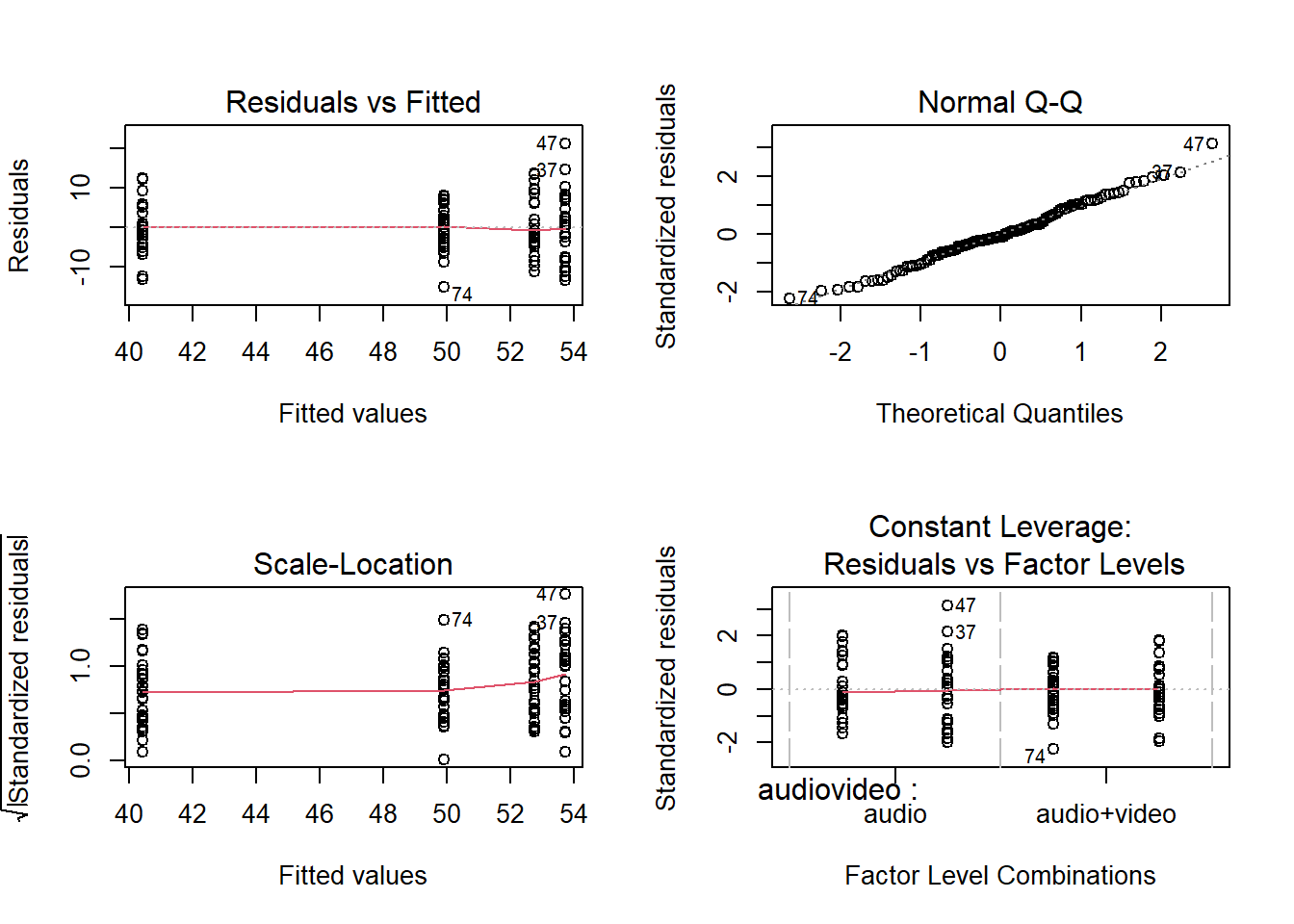
Step 2 is to run your model(s) of interest to answer your research question, and make sure that the data meet the assumptions of your chosen test.
- Conduct a two-way ANOVA
- Check assumptions
The third and final step(s) somewhat depends on the outcomes of step 2. Here, you may need to consider conducting further analyses before writing up / describing your results in relation to the research question.
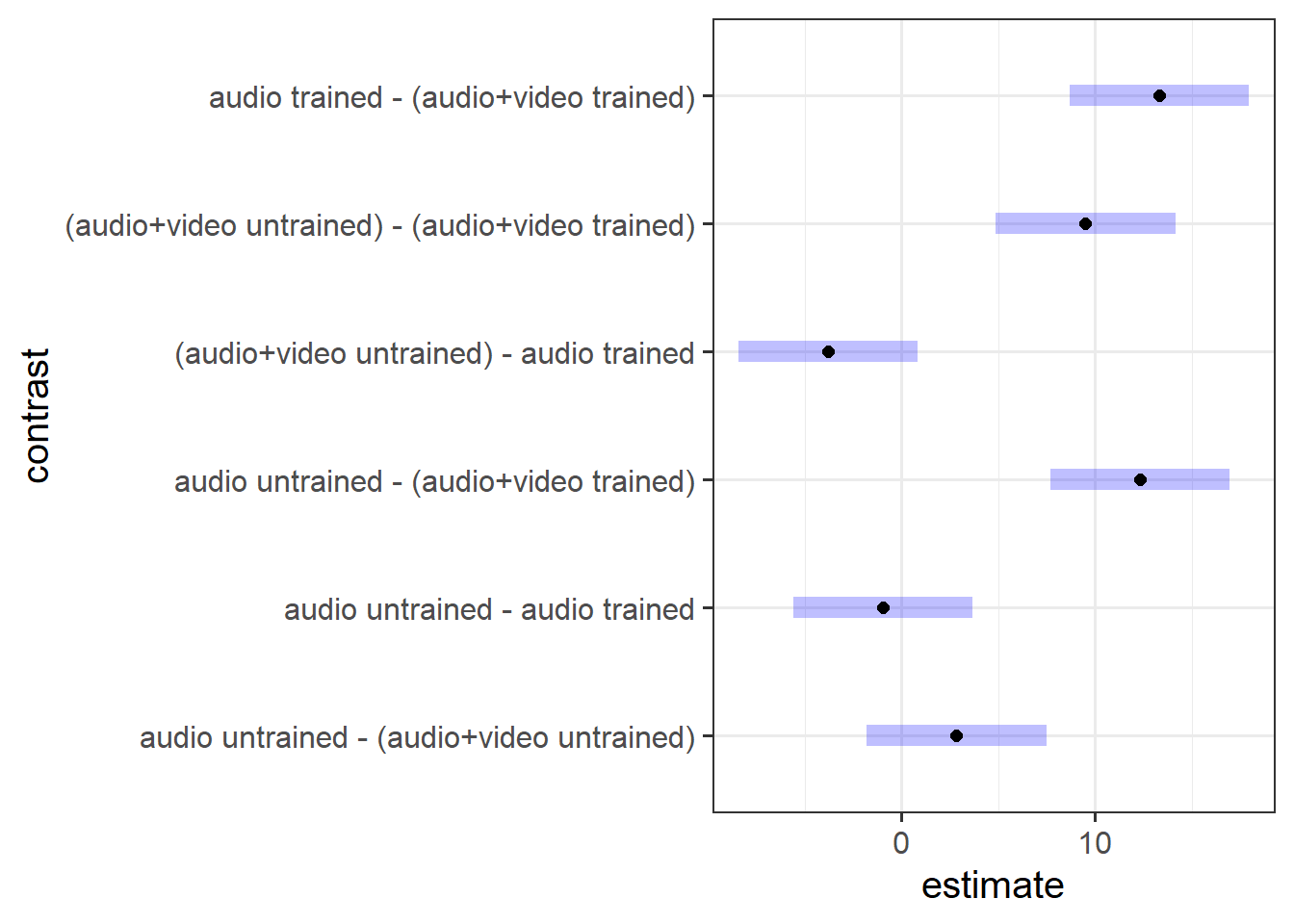
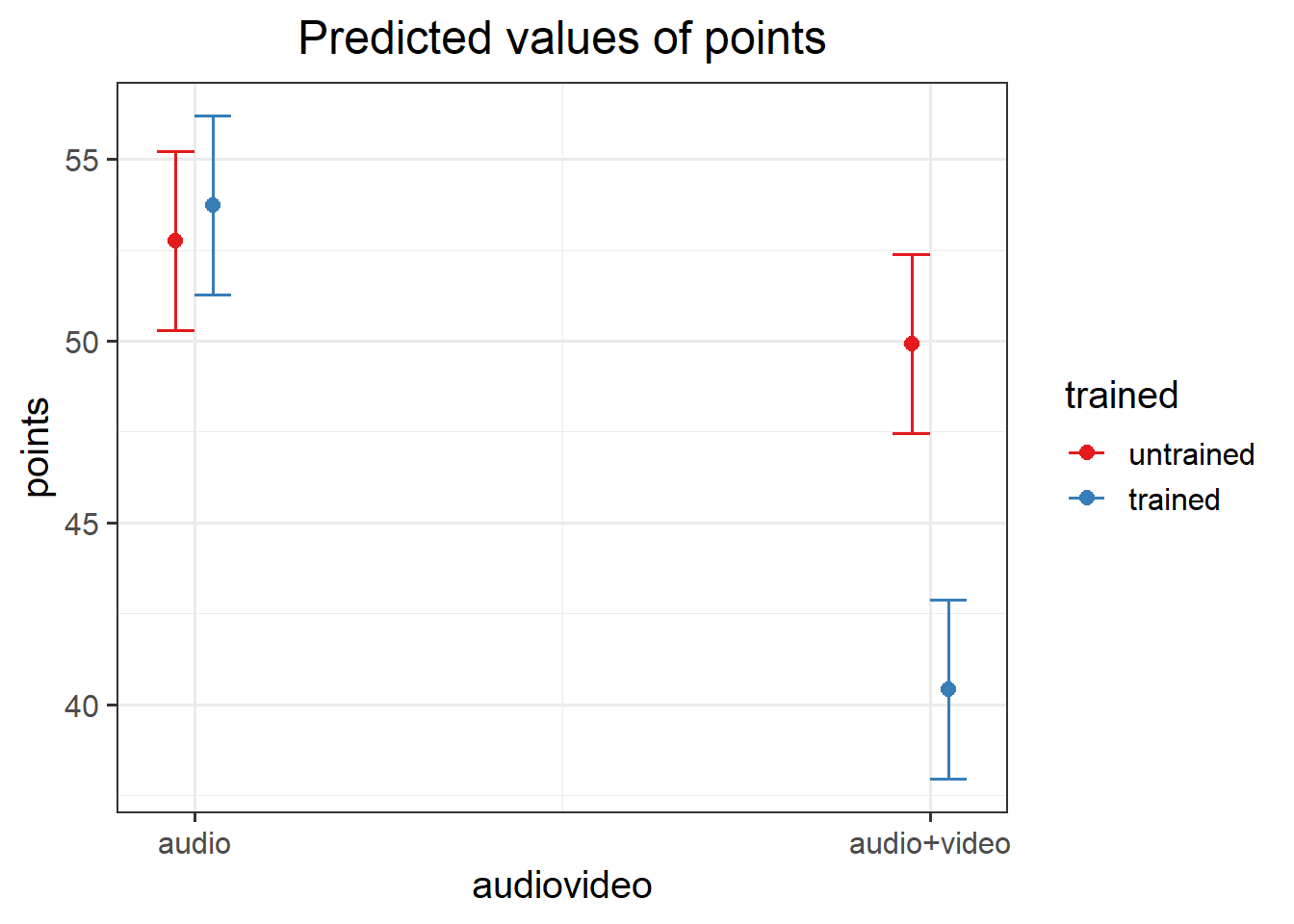
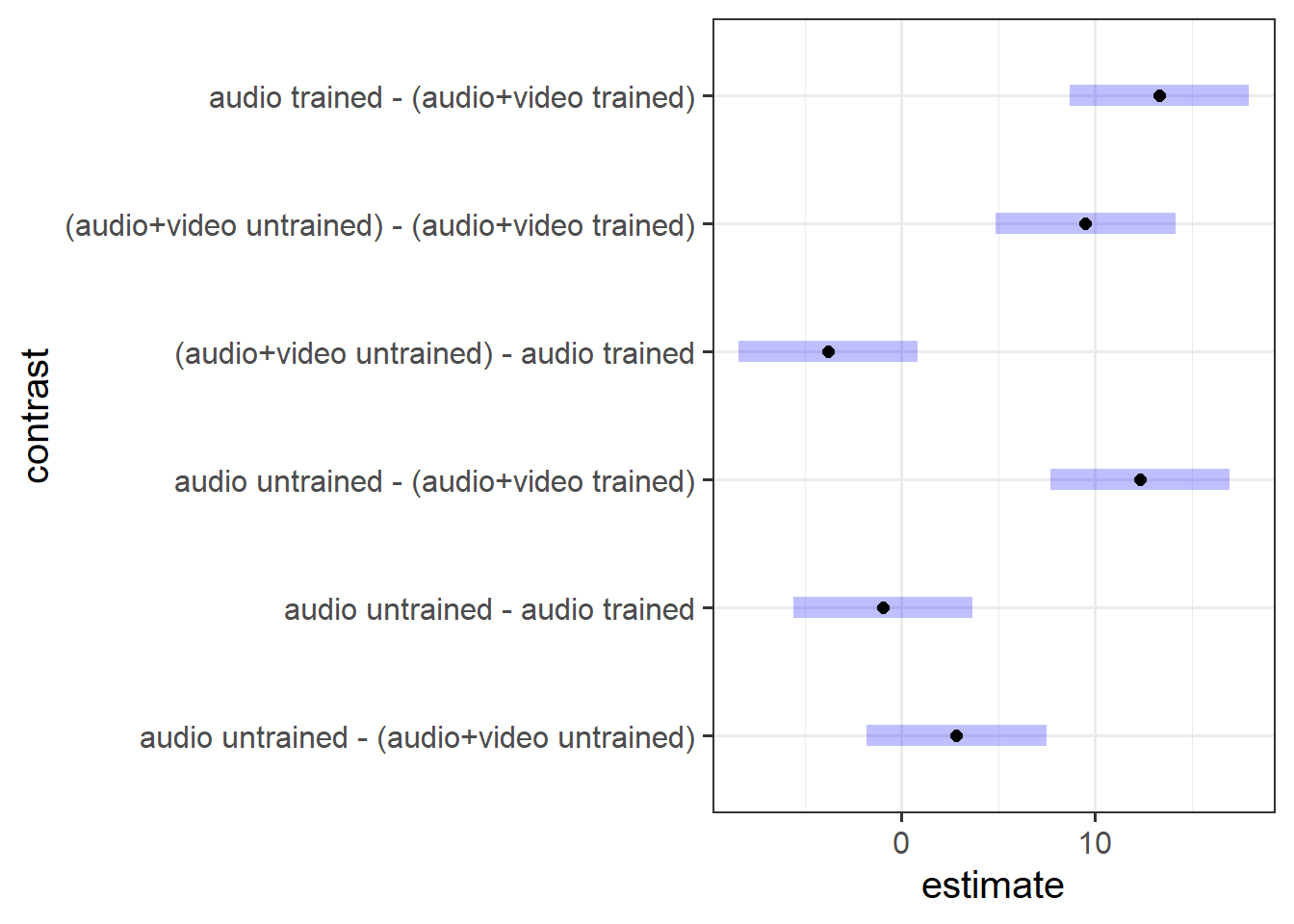
- Perform a pairwise comparison of the mean accuracy (as measured by points accrued) across the 2×2 factorial design, making sure to adjust for multiple comparisons by the method of your choice.
- Interpret your results in relation to the research question (remember you can refer to your previous plotting when doing this!).
Analysis Strategy
Results
Discussion
what defines a ‘family’ of tests is debateable.↩︎