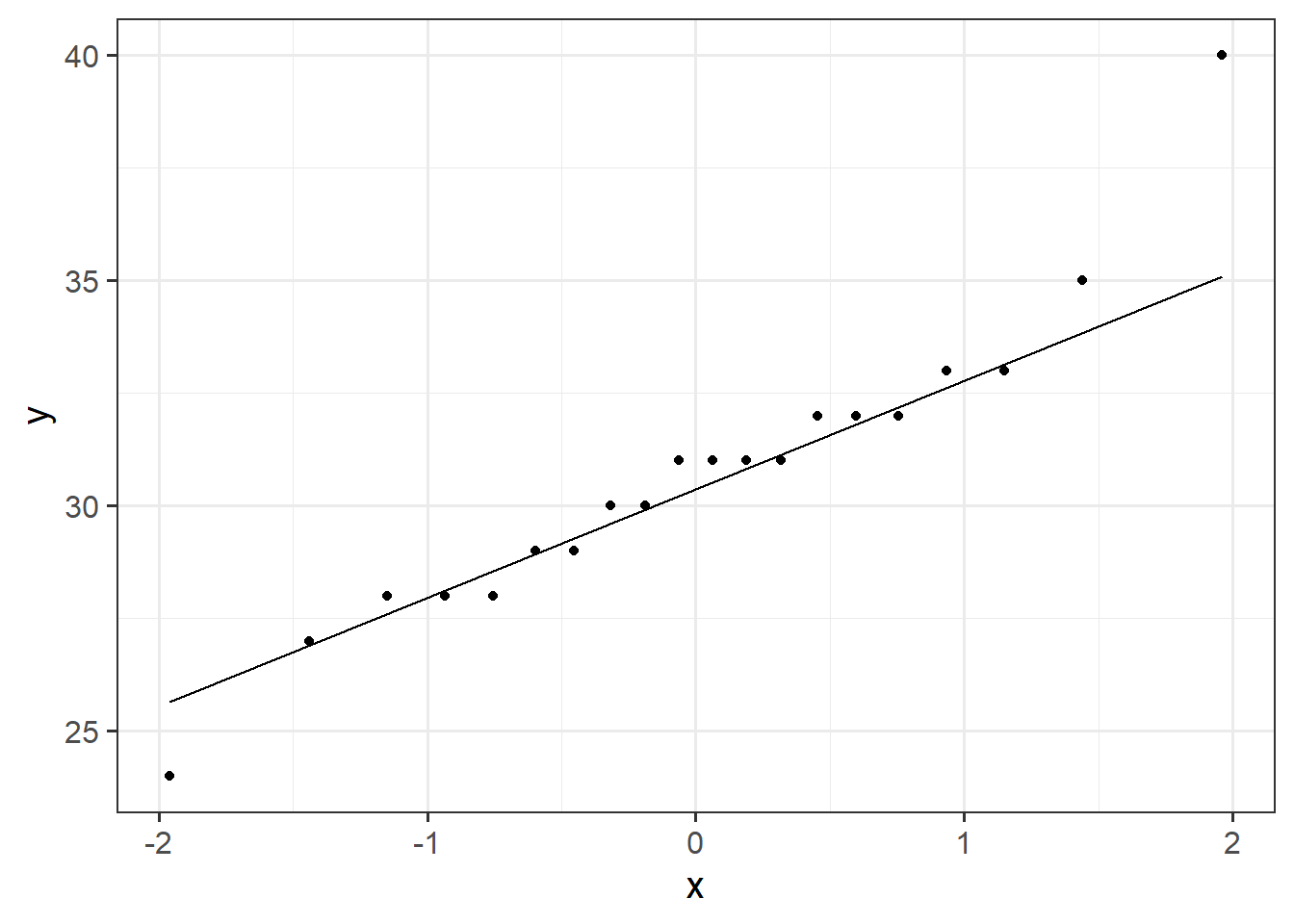pass_scores <- read_csv("https://uoepsy.github.io/data/pass_scores.csv") Week 3 Exercises: T-tests
Procrastination Scores
Research Question Do Edinburgh University students report endorsing procrastination less than the norm?
The Procrastination Assessment Scale for Students (PASS) was designed to assess how individuals approach decision situations, specifically the tendency of individuals to postpone decisions (see Solomon & Rothblum, 1984). The PASS assesses the prevalence of procrastination in six areas: writing a paper; studying for an exam; keeping up with reading; administrative tasks; attending meetings; and performing general tasks. For a measure of total endorsement of procrastination, responses to 18 questions (each measured on a 1-5 scale) are summed together, providing a single score for each participant (range 0 to 90). The mean score from Solomon & Rothblum, 1984 was 33.
A student administers the PASS to 20 students from Edinburgh University.
The data are available at https://uoepsy.github.io/data/pass_scores.csv.
- Read in the data
- Calculate some relevant descriptive statistics
- Check the assumptions that we will be concerned with for a one-sample test of whether the mean PASS scores is less than 33.
Our test here is going to be have the following hypotheses:
- Null: mean PASS score in Edinburgh Uni students is \(\geq 33\)
- Alternative: mean PASS score in Edinburgh Uni students is \(< 33\)
Read in the data and manually calculate the relevant test statistic.
Note, we’re doing this manually right now as it’s a useful learning process. In later questions we will switch to the easy way!
Hints:
- you can see the manual calculation of a one sample t-test in 3B #one-sample-t-test.
- The relevant formula is:
\[ \begin{align} & t = \frac{\bar x - \mu_0}{\frac{s}{\sqrt{n}}} \\ \qquad \\ & \text{Where:} \\ & \bar x : \text{mean of PASS in our sample} \\ & \mu_0 : \text{hypothesised mean score of 33} \\ & s : \text{standard deviation of PASS in our sample} \\ & n : \text{number of observations} \end{align} \]
Using the test statistic calculated in question 1, compute the p-value.
Hint:
- this will be needing the
pt()function. - the degrees of freedom is \(n-1\) (we used one up by estimating the mean).
- The test we are performing is against the null hypothesis that the mean is \(\geq 33\). Our t-statistic is in the broad sense calculated as “mean minus 33”, so negative numbers mean we have a mean lower than 33. These are the instances that we will reject the null hypothesis - if we get a test statistic very low. So we want the lower.tail of the distribution for our p-value.
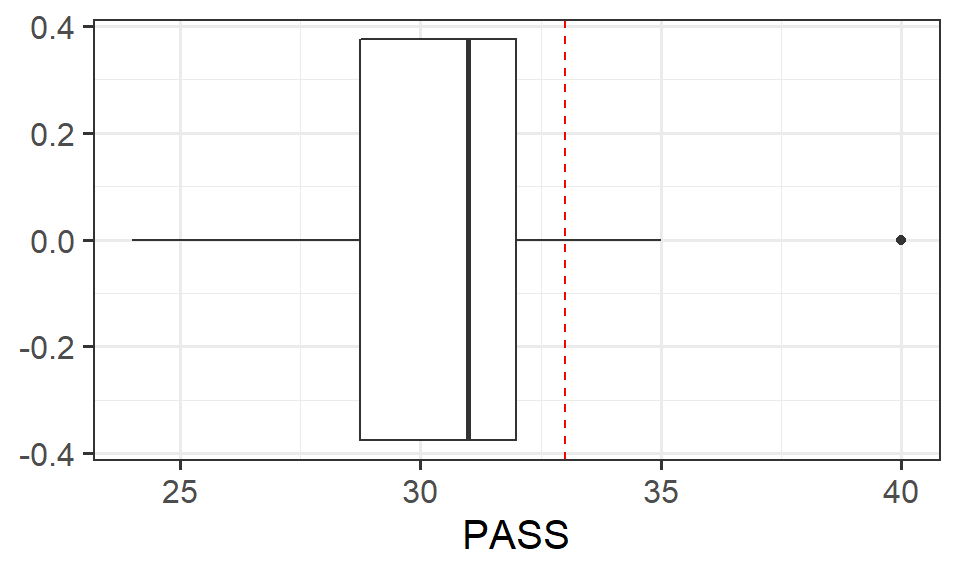
Now using the t.test() function, conduct the same test. Check that the numbers match with questions 1 and 2, and create a visualisation to illustrate any results.
Hint: Check out the help page for t.test() - there is an argument in the function that allows us to easily change between whether our alternative hypothesis is “less than”, “greater than” or “not equal to”.
Write up the results.
Hints: There are some quick example write-ups for each test in 3B #basic-tests
Heights
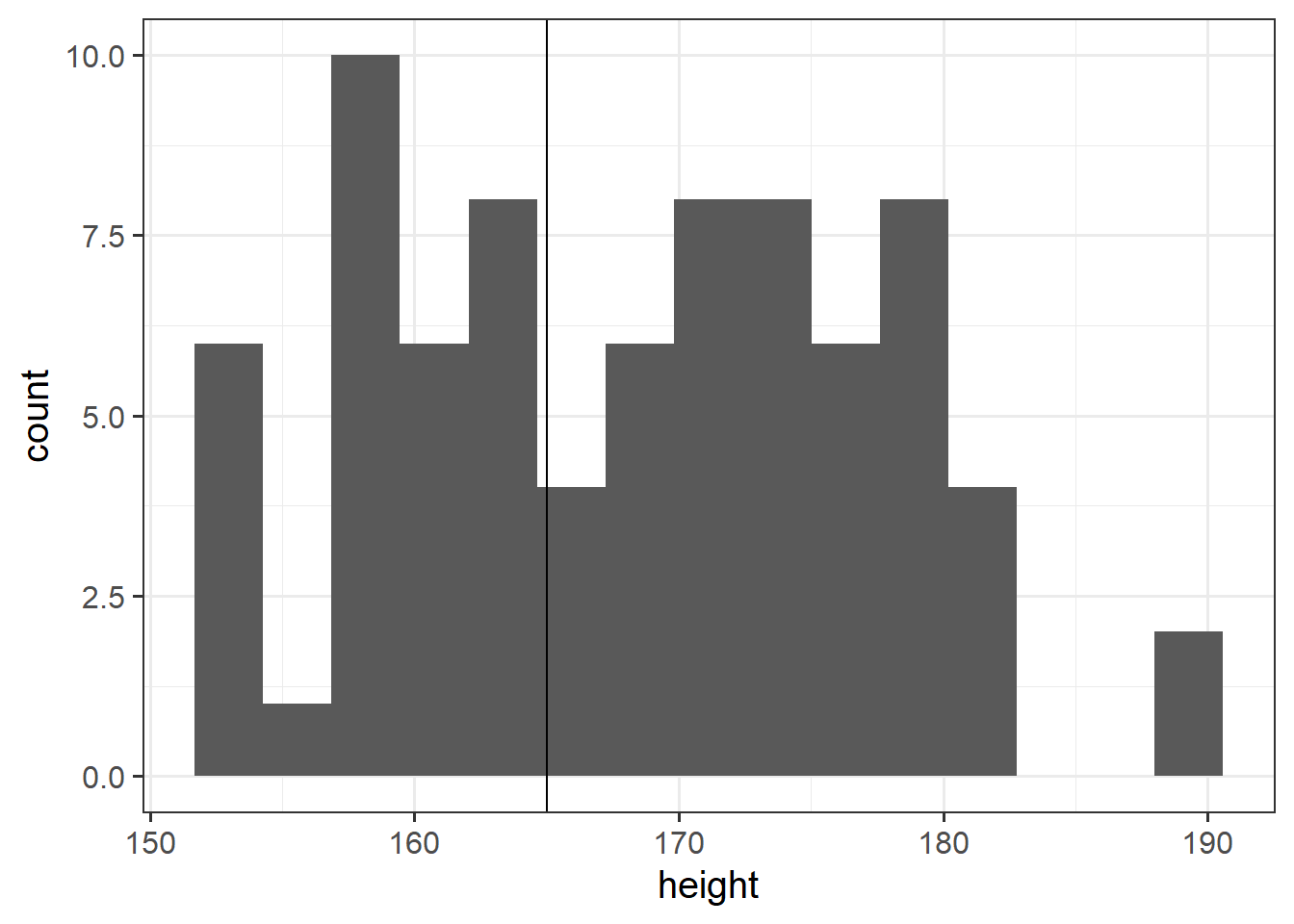
Research Question Is the average height of students taking the USMR statistic course in Psychology at Edinburgh University in 2021/2022 is different from 165cm?

The data for students from all psychology statistics courses last year and USMR this year, are available at https://uoepsy.github.io/data/surveydata_allcourse22.csv.
No more manual calculations of test statistics and p-values for this week.
Conduct a one sample \(t\)-test to evaluate whether the average height of students taking the USMR courses in Psychology at Edinburgh University in 2022/23 is different from 165cm.
Hints:
- This is real data, and real data is rarely normal! If you conduct a Shapiro-Wilk test, you may well find \(p<.05\) and conclude that your data is not normal.
So what do we do if a test indicates our assumptions are violated?
Well, we should bear a couple of things in mind.- A decision rule such as \(p<.05\) on Shapiro-Wilk test creates very dichotomous thinking for something which is in reality not black and white. Real life distributions are not either normal or non-normal. Plot the data, and make a judgement!
- As it happens, the t-test is actually reasonably robust against slight deviations from normality! Plot your data and make a judgement!
- The deeper you get into statistics, the more you discover that it is not simply a case of following step-by-step rules:
- A decision rule such as \(p<.05\) on Shapiro-Wilk test creates very dichotomous thinking for something which is in reality not black and white. Real life distributions are not either normal or non-normal. Plot the data, and make a judgement!

Names and Tips
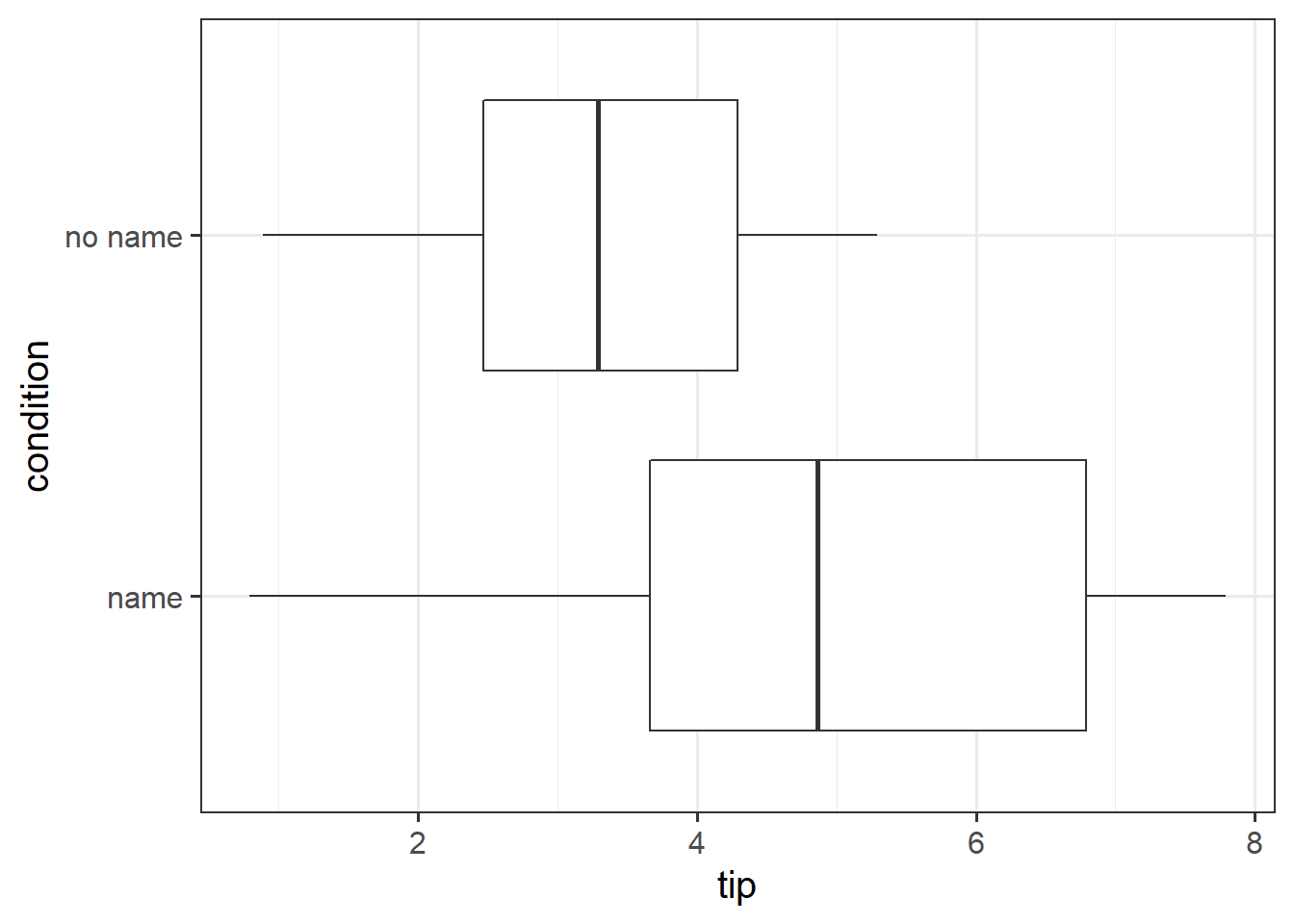
Research Question Can a server earn higher tips simply by introducing themselves by name when greeting customers?
Researchers investigated the effect of a server introducing herself by name on restaurant tipping. The study involved forty, 2-person parties eating a $23.21 fixed-price buffet Sunday brunch at Charley Brown’s Restaurant in Huntington Beach, California, on April 10 and 17, 1988. Each two-person party was randomly assigned by the waitress to either a name or a no name introduction condition using a random mechanism. The waitress kept track of the two-person party condition and how much the party paid at the end of the meal.
The data are available at https://uoepsy.github.io/data/gerritysim.csv. (This is a simulated example based on Garrity and Degelman (1990))
Conduct an independent samples \(t\)-test to assess whether higher tips are earned when the server introduces themselves by name, in comparison to when they do not.
Hints:
- We’ll want to check the normality (either visually or with a test) of the variable of interest for each group.
- Some researchers suggest using the Welch t-test by default. This means you can relax the assumption of equal variances in the groups. If you want to test whether two variances are equal, try the
var.test()function.
Optional Extras
Here are a few extra questions for you to practice performing tests and making plots:
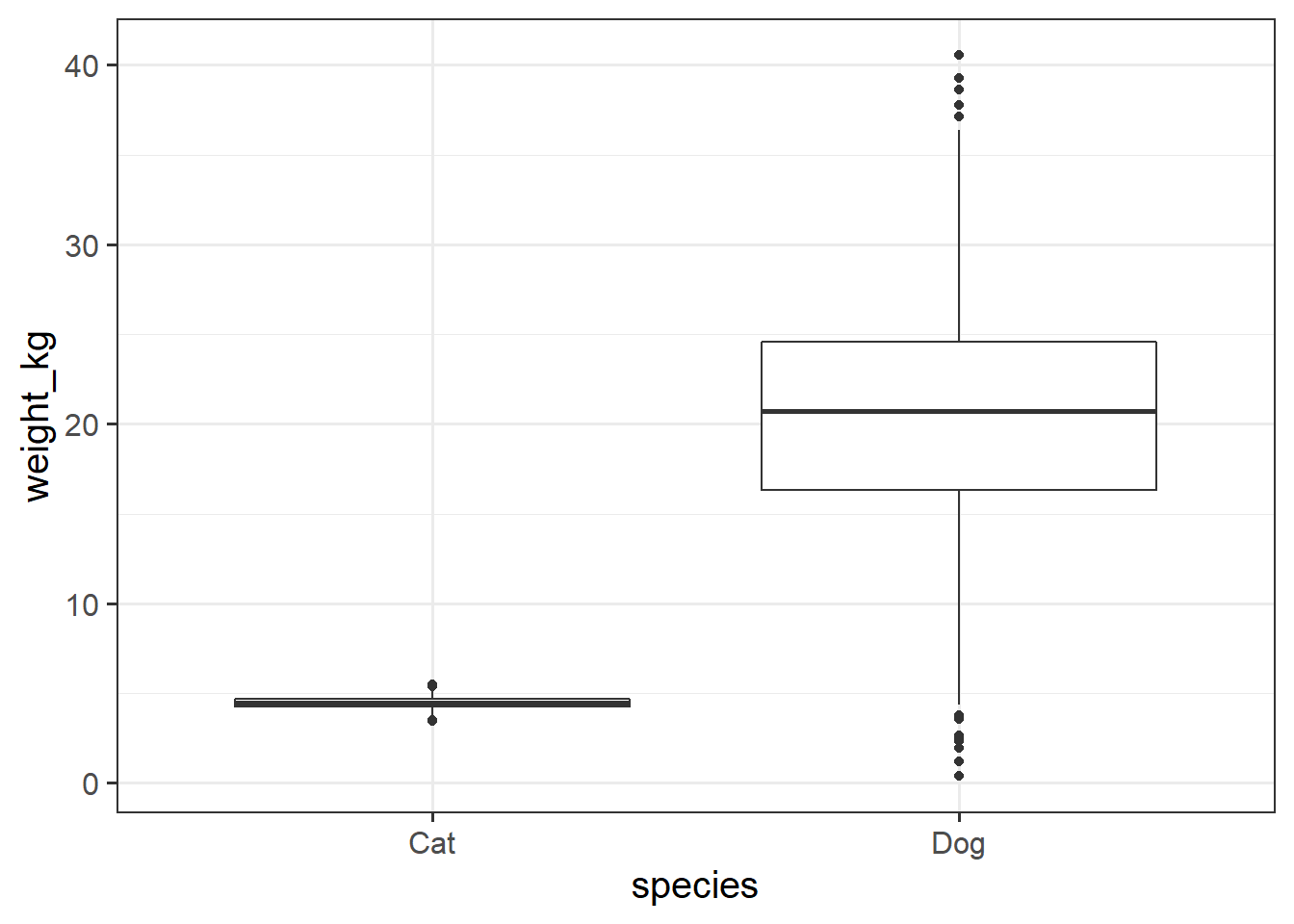
- Are dogs heavier on average than cats?
- Data are at https://uoepsy.github.io/data/pets_seattle.csv
- Remember from week 1 - not everything in that data is either a cat or a dog!
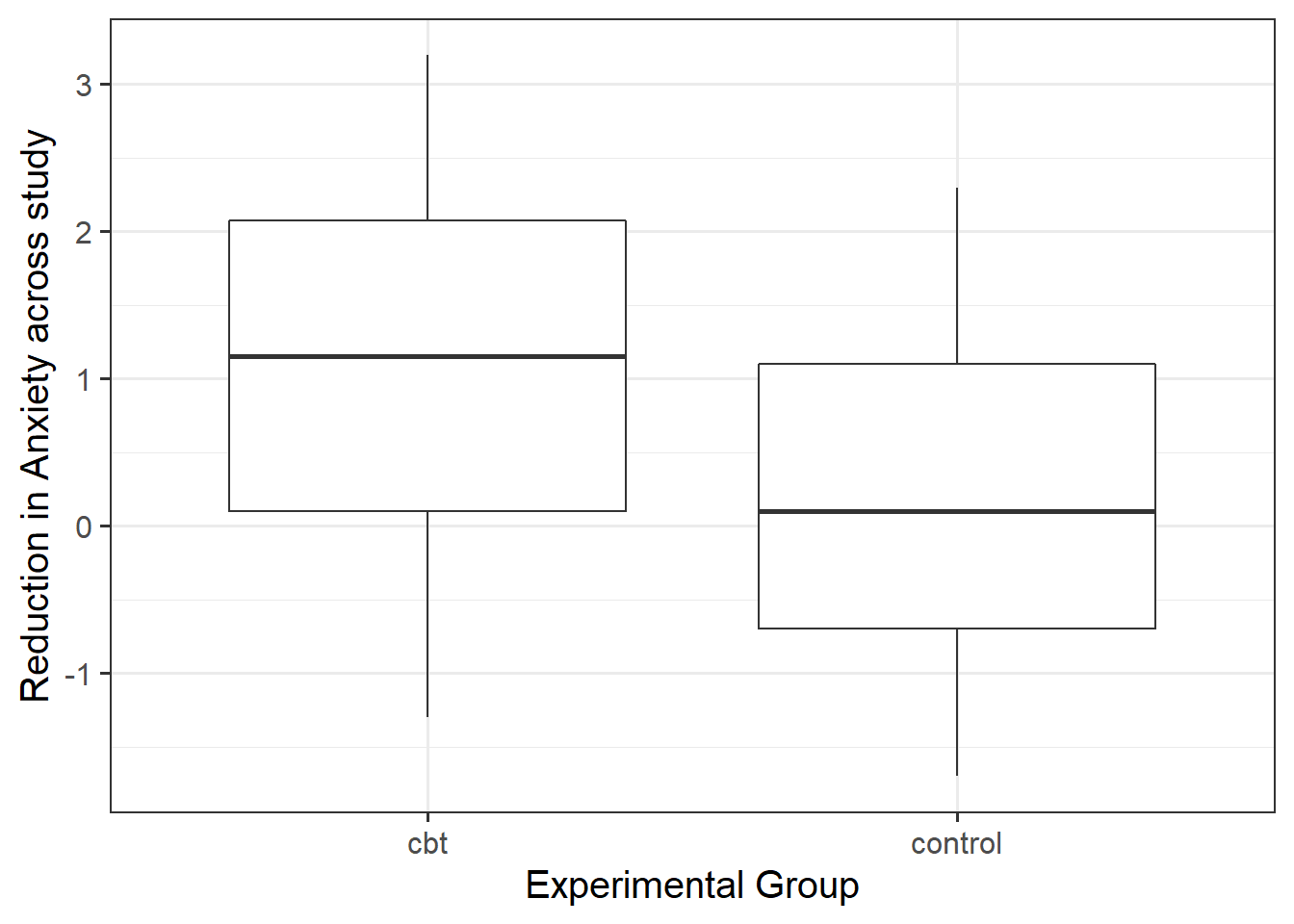
- Is taking part in a cognitive behavioural therapy (CBT) based programme associated with a greater reduction, on average, in anxiety scores in comparison to a Control group?
- Data are at https://uoepsy.github.io/data/dapr1_2122_report_data.csv. The dataset contains information on each person in an organisation, recording their professional role (management vs employee), whether they are allocated into the CBT programme or not (control vs cbt), and scores on anxiety at both the start and the end of the study period.
- you might have to make a new variable in order to test the research question.