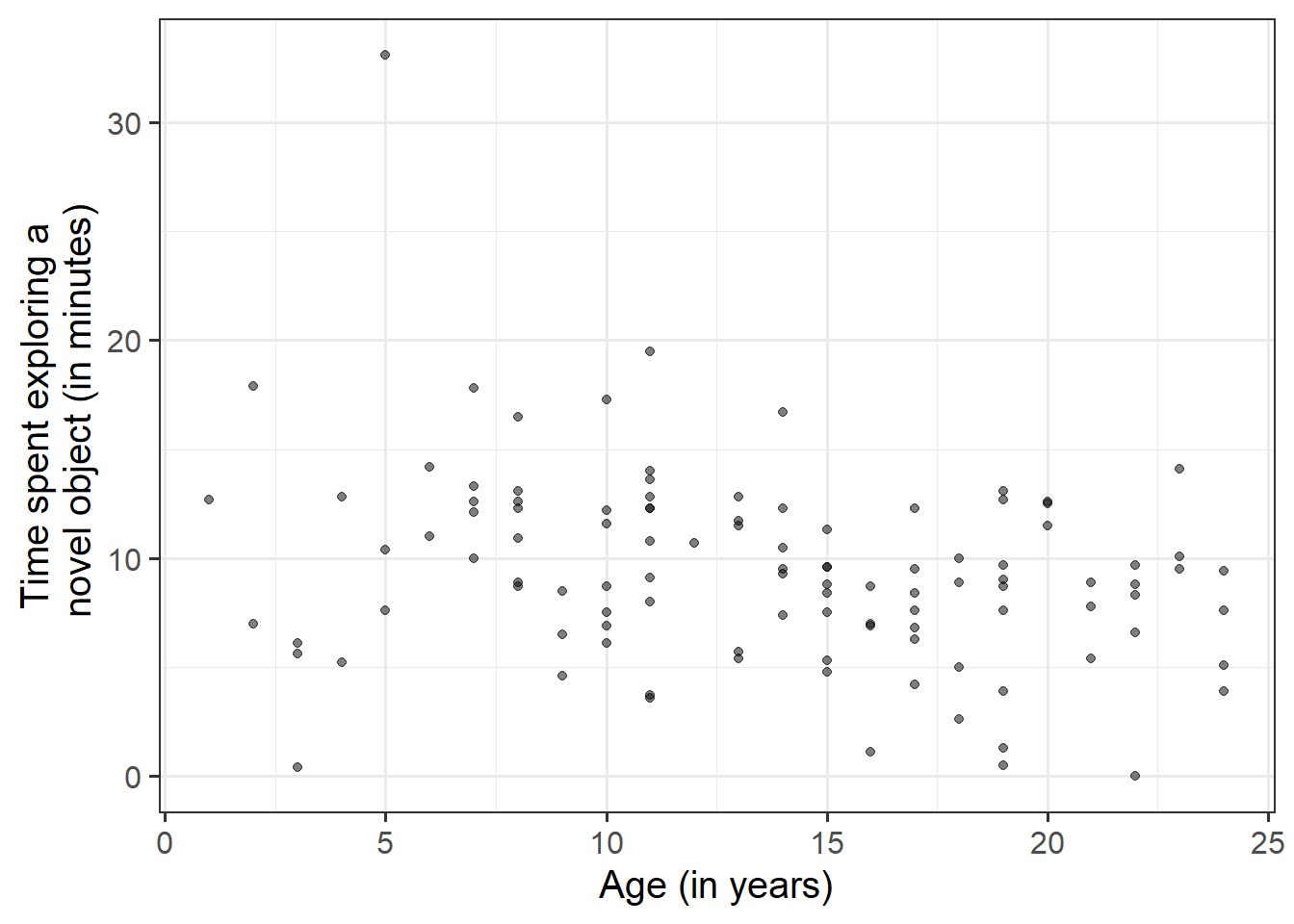| Item | Statement |
|---|---|
| Item_1 | I often felt an inability to concentrate |
| Item_2 | I frequently forgot things |
| Item_3 | I found thinking clearly required a lot of effort |
| Item_4 | I often felt happy |
| Item_5 | I had lots of energy |
| Item_6 | I worked efficiently |
| Item_7 | I often felt irritable |
| Item_8 | I often felt stressed |
| Item_9 | I often felt sleepy |
| Item_10 | I often felt fatigued |
Exercises: Covariance, Correlation & Linear Regression
Covariance & Correlation
Go to http://guessthecorrelation.com/ and play the “guess the correlation” game for a little while to get an idea of what different strengths and directions of \(r\) can look like.
Sleepy time
Data: Sleep levels and daytime functioning
A researcher is interested in the relationship between hours slept per night and self-rated effects of sleep on daytime functioning. She recruited 50 healthy adults, and collected data on the Total Sleep Time (TST) over the course of a seven day period via sleep-tracking devices.
At the end of the seven day period, participants completed a Daytime Functioning (DTF) questionnaire. This involved participants rating their agreement with ten statements (see Table 1). Agreement was measured on a scale from 1-5. An overall score of daytime functioning can be calculated by:
- reversing the scores for items 4,5 and 6 (because those items reflect agreement with positive statements, whereas the other ones are agreement with negative statement);
- summing the scores on each item; and
- subtracting the sum score from 50 (the max possible score). This will make higher scores reflect better perceived daytime functioning.
The data is available at https://uoepsy.github.io/data/sleepdtf.csv.
Load the required libraries (probably just tidyverse for now), and read in the data.
Calculate the overall daytime functioning score, following the criteria outlined above, and make this a new column in your dataset.
To reverse items 4, 5 and 6, we we need to make all the scores of 1 become 5, scores of 2 become 4, and so on… What number satisfies all of these equations: ? - 5 = 1, ? - 4 = 2, ? - 3 = 3?
To quickly sum across rows, you might find the rowSums() function useful (you don’t have to use it though)
If my items were in columns between 4 to 15:
dataframe$sumscore = rowSums(dataframe[, 4:15])
Calculate the correlation between the total sleep time (TST) and the overall daytime functioning score calculated in the previous question.
Conduct a test to establish the probability of observing a correlation this strong in a sample of this size assuming the true correlation to be 0.
Write a sentence or two summarising the results.
You can do this all with one function, see 5A #correlation-test.
Think about this relationship in terms of causation.
Claim: Less sleep causes poorer daytime functioning.
Why might it be inappropriate to make the claim above based on these data alone? Think about what sort of study could provide stronger evidence for such a claim.
- comparison groups.
- random allocation.
- measures of daytime functioning.
- measures of sleep time.
- other (unmeasured) explanatory variables.
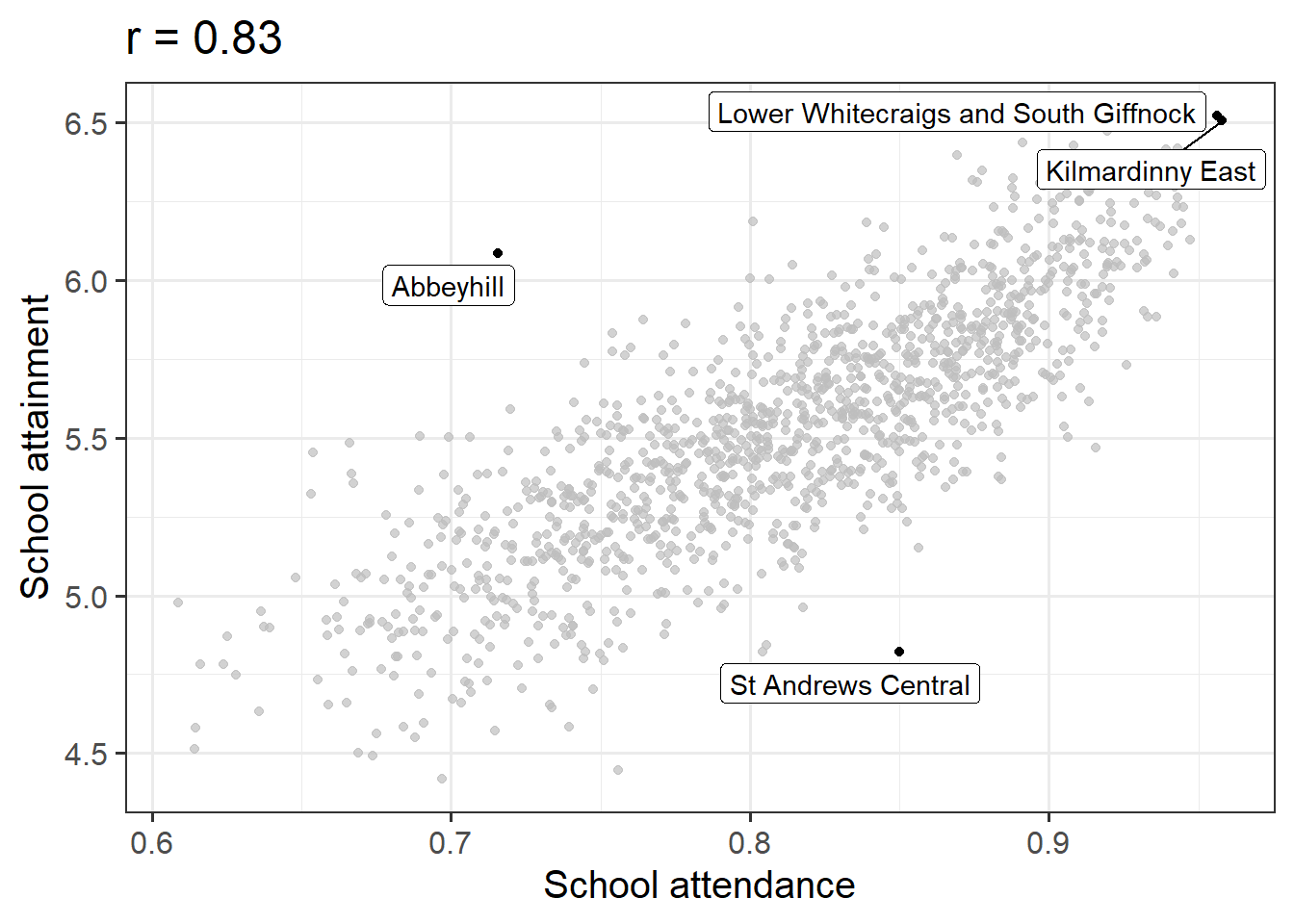
Attendance and Attainment
Data: Education SIMD Indicators
The Scottish Government regularly collates data across a wide range of societal, geographic, and health indicators for every “datazone” (small area) in Scotland.
The dataset at https://uoepsy.github.io/data/simd20_educ.csv contains some of the education indicators (see Table 2).
| variable | description |
|---|---|
| intermediate_zone | Areas of scotland containing populations of between 2.5k-6k household residents |
| attendance | Average School pupil attendance |
| attainment | Average attainment score of School leavers (based on Scottish Credit and Qualifications Framework (SCQF)) |
| university | Proportion of 17-21 year olds entering university |
Conduct a test of whether there is a correlation between school attendance and school attainment in Scotland.
Present and write up the results.
The readings have not included an example write-up for you to follow. Try to follow the logic of those for t-tests and \(\chi^2\)-tests.
- describe the relevant data
- explain what test was conducted and why
- present the relevant statistic, degrees of freedom (if applicable), statement on p-value, etc.
- state the conclusion.
Be careful figuring out how many observations your test is conducted on. cor.test() includes only the complete observations.
Simple Linear Regression
Monkey Exploration
Data: monkeyexplorers.csv
Liu, Hajnosz & Li (2023)1 have conducted a study on monkeys! They were interested in whether younger monkeys tend to be more inquisitive about new things than older monkeys do. They sampled 108 monkeys ranging from 1 to 24 years old. Each monkey was given a novel object, the researchers recorded the time (in minutes) that each monkey spent exploring the object.
For this week, we’re going to be investigating the research question:
Do older monkeys spend more/less time exploring novel objects?
The data is available at https://uoepsy.github.io/data/monkeyexplorers.csv and contains the variables described in Table 3
| variable | description |
|---|---|
| name | Monkey Name |
| age | Age of monkey in years |
| exploration_time | Time (in minutes) spent exploring the object |
For this week, we’re going to be investigating the following research question:
Do older monkeys spend more/less time exploring novel objects?
Read in the data to your R session, then visualise and describe the marginal distributions of those variables which are of interest to us. These are the distribution of each variable (time spent exploring, and monkey age) without reference to the values of the other variables.
- You could use, for example,
geom_density()for a density plot orgeom_histogram()for a histogram. - Look at the shape, center and spread of the distribution. Is it symmetric or skewed? Is it unimodal or bimodal?
- Do you notice any extreme observations?
After we’ve looked at the marginal distributions of the variables of interest in the analysis, we typically move on to examining relationships between the variables.
Visualise and describe the relationship between age and exploration-time among the monkeys in the sample.
Think about:
- Direction of association
- Form of association (can it be summarised well with a straight line?)
- Strength of association (how closely do points fall to a recognizable pattern such as a line?)
- Unusual observations that do not fit the pattern of the rest of the observations and which are worth examining in more detail.
Using the lm() function, fit a linear model to the sample data, in which time that monkeys spend exploring novel objects is explained by age. Assign it to a name to store it in your environment.
You can see how to fit linear models in R using lm() in 5B #fitting-linear-models-in-r
Interpret the estimated intercept and slope in the context of the question of interest.
We saw how to extract lots of information on our model using summary() (see 5B #model-summary), but there are lots of other functions too.
If we called our linear model object “model1” in the environment, then we can use:
- type
model1, i.e. simply invoke the name of the fitted model; - type
model1$coefficients; - use the
coef(model1)function; - use the
coefficients(model1)function; - use the
summary(model1)$coefficientsto extract just that part of the summary.
Test the hypothesis that the population slope is zero — that is, that there is no linear association between exploration time and age in the population.
You don’t need to do anything for this, you can find all the necessary information in summary() of your model.
See 5B #inference-for-regression-coefficients.
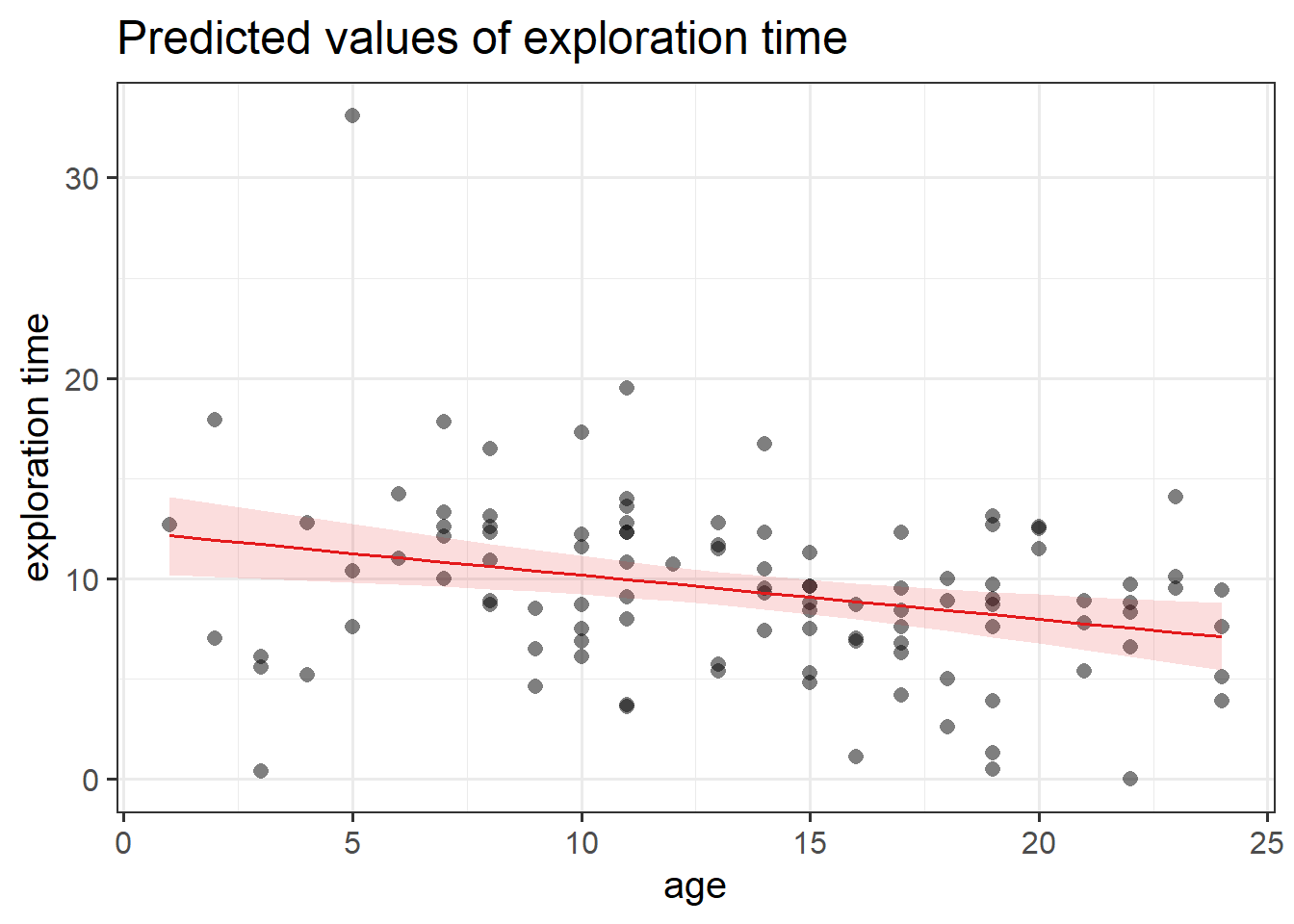
Create a visualisation of the estimated association between age and exploration time.
There are lots of ways to do this. Check 5B #example, which shows an example of using the sjPlot package to create a plot.
Consider the following:
- In fitting a linear regression model, we make the assumption that the errors around the line are normally distributed around zero (this is the \(\epsilon \sim N(0, \sigma)\) bit.)
- About 95% of values from a normal distribution fall within two standard deviations of the centre.
We can obtain the estimated standard deviation of the errors (\(\hat \sigma\)) from the fitted model using sigma() and giving it the name of our model.
What does this tell us?
See 5B #the-error.
Compute the model-predicted exploration-time for a monkey that is 1 year old.
Given that you know the intercept and slope, you can calculate this algebraically. However, try to also use the predict() function (see 5B #model-predictions).
Influential Monkeys
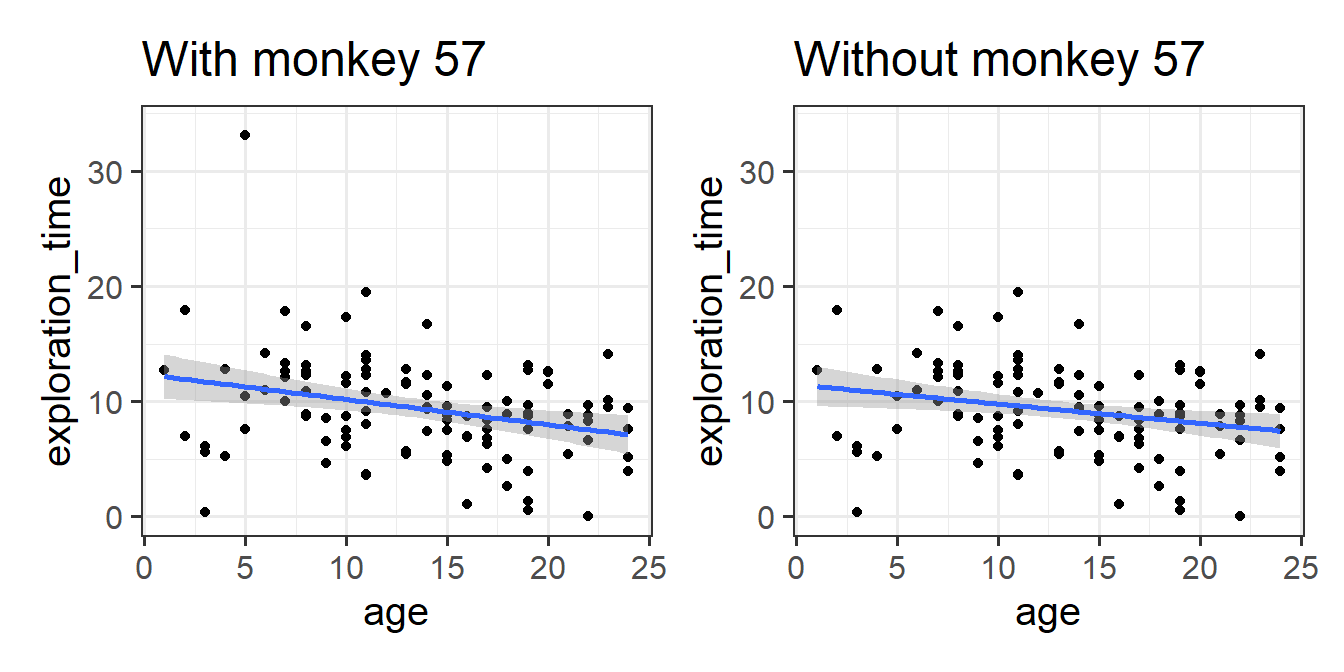
Take a look at the assumption plots (see 5B #assumptions) for your model.
- The trick to looking at assumption plots in linear regression is to look for “things that don’t look random”.
- As well as looking for patterns, these plots can also higlight individual datapoints that might be skewing the results. Can you figure out if there are any unusual monkeys in our dataset? Can you re-fit the model without that monkey? When you do so, do your conclusions change?
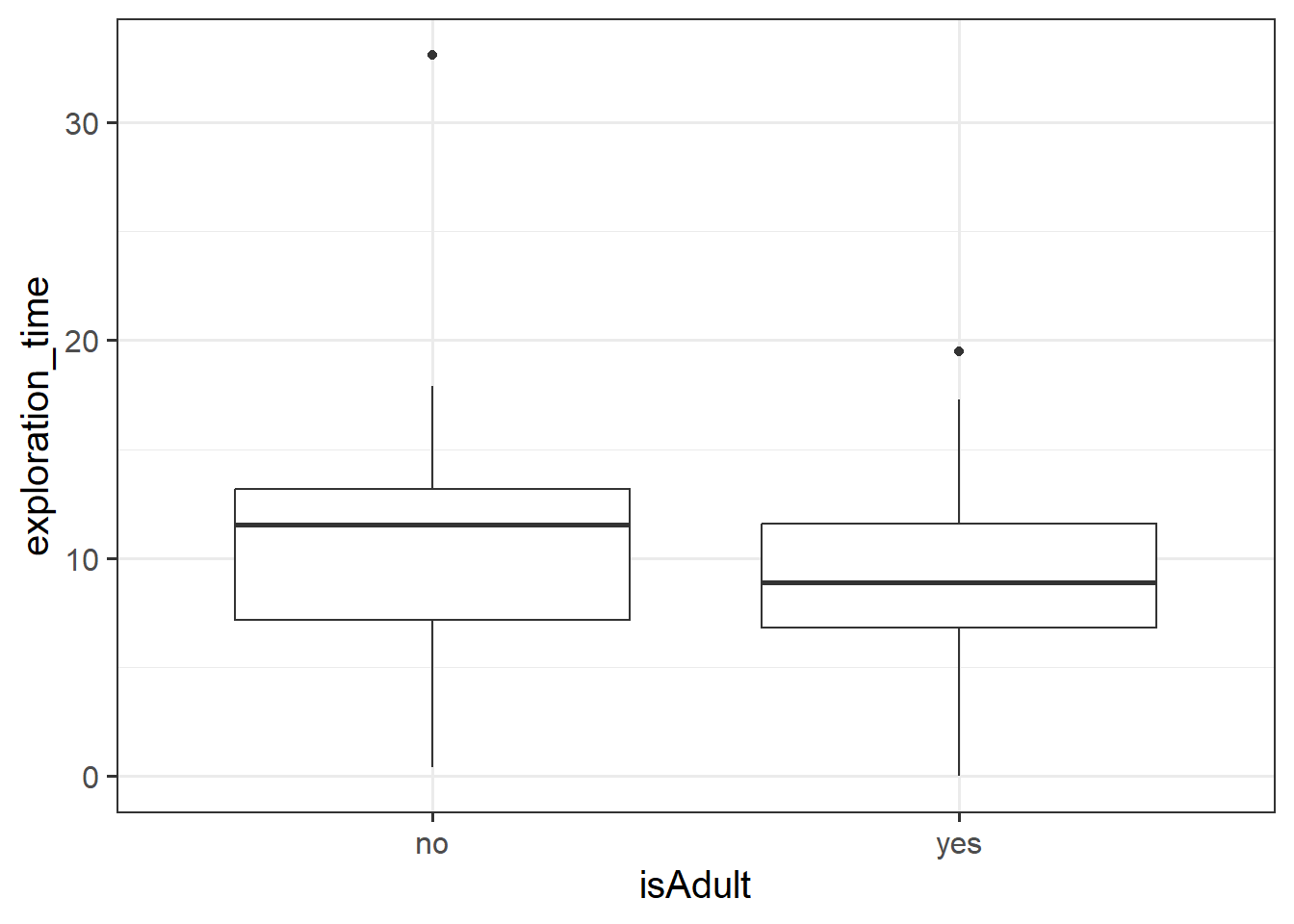
Monkey Exploration in Adulthood
Let’s suppose instead of having measured monkeys’ ages in years, researchers simply recorded whether each monkey was an adult or a juvenile (both Capuchins and Rhesus Macaques reach adulthood at 8 years old).
The code below creates a this new variable for us:
monkeyexp <- monkeyexp |>
mutate(
isAdult = ifelse(age >= 8, "yes","no")
)Fit the following model, and interpret the coefficients.
\[ \text{ExplorationTime} = b_0 + b_1 \cdot \text{isAdult} + \varepsilon \]
For help interpreting the coefficients, see 5B #binary-predictors.
We’ve actually already seen a way to analyse questions of this sort (“is the average exploration-time different between juveniles and adults?”)
Run the following t-test, and consider the statistic, p value etc. How does it compare to the model in the previous question?
t.test(exploration_time ~ isAdult, data = monkeyexp, var.equal = TRUE)
Extra! A team task!
The data from this year’s survey that you filled out in the first couple of weeks is available at https://uoepsy.github.io/data/usmr2023.csv.
- Step 1: Using the USMR 2023 survey data, make either the prettiest plot you can or the ugliest plot you can .
- Step 2: Submit your plot! Go to https://forms.office.com/e/KiGevk6xH3 and upload your plot and the code used to create it (save the plot via the plot window in R/take a screenshot/put it in a .Rmd and knit/whatever gets the picture uploaded!)
All plots submitted by end-of-day on 25th October will be rated by a number of non-USMR staff, and prizes made of chocolate will be awarded to the team with the prettiest plot and to the team with the ugliest plot.
Footnotes
Not a real study!↩︎