| variable | description |
|---|---|
| Diagnosis | Diagnosis classifies the three types of individuals: 1 = Amnesic patients, 2 = Huntingtons patients, and 3 = Control group of individuals with no known neurological disorder |
| Task | Task tells us to which one of two tasks each study participant was randomly assigned to: 1 = Grammar (which consists of classifying letter sequences as either following or not following grammatical rules), and 2 = Recognition (which consists of recognising particular stimuli as stimuli that have previously been presented during the task) |
| Y | Score |
Interactions III: Cat x Cat
Learning Objectives
At the end of this lab, you will:
- Understand the concept of an interaction
- Be able to interpret a categorical \(\times\) categorical interaction
- Be able to visualize and probe interactions
What You Need
Required R Packages
Remember to load all packages within a code chunk at the start of your RMarkdown file using library(). If you do not have a package and need to install, do so within the console using install.packages(" "). For further guidance on installing/updating packages, see Section C here.
For this lab, you will need to load the following package(s):
- tidyverse
- psych
- sjPlot
- kableExtra
- sandwich
- interactions
Presenting Results
All results should be presented following APA guidelines. If you need a reminder on how to hide code, format tables/plots, etc., make sure to review the rmd bootcamp.
The example write-up sections included as part of the solutions are not perfect - they instead should give you a good example of what information you should include and how to structure this. Note that you must not copy any of the write-ups included below for future reports - if you do, you will be committing plagiarism, and this type of academic misconduct is taken very seriously by the University. You can find out more here.
Lab Data
You can download the data required for this lab here or read it in via this link https://uoepsy.github.io/data/cognitive_experiment_3_by_2.csv
Study Overview
Research Question
Are there differences in types of memory deficits for those experiencing different cognitive impairment(s)?
Setup
- Create a new RMarkdown file
- Load the required package(s)
- Read the cognitive_experiment_3_by_2 dataset into R, assigning it to an object named
cog
Exercises
Study & Analysis Plan Overview
Examine the dataset, and perform any necessary and appropriate data management steps.
Choose appropriate reference levels for the Diagnosis and Task variables.
Read the Study Overview codebook carefully.
Provide a brief overview of the study design and data, before detailing your analysis plan to address the research question.
- Give the reader some background on the context of the study
- State what type of analysis you will conduct in order to address the research question
- Specify the model to be fitted to address the research question (note that you will need to specify the reference level of your categorical variables)
- Specify your chosen significance (\(\alpha\)) level
- State your hypotheses
Much of the information required can be found in the Study Overview codebook.
Descriptive Statistics & Visualisations
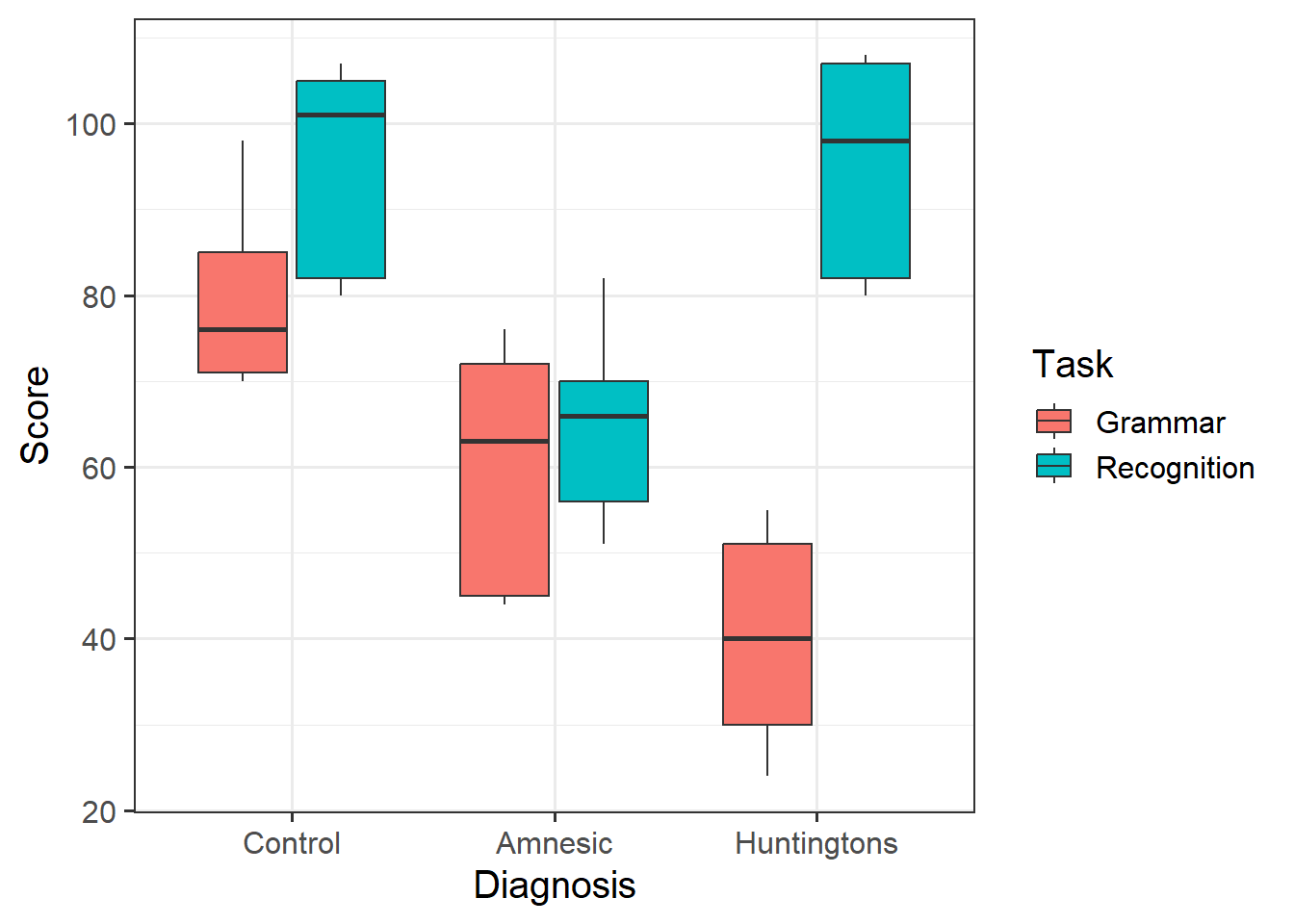
Provide a table of descriptive statistics and visualise your data.
Remember to interpret your plot in the context of the study.
- For your table of descriptive statistics, both the
group_by()andsummarise()functions will come in handy here. - Recall that when visualising categorical variables,
geom_boxplot()may be most appropriate to use.
Model Fitting & Interpretation
Fit the specified model using lm(), and store the model in an object named “cog_mdl”.
R?
Fortunately, R computes the dummy variables for us! Thus, each row in the summary() output of the model will correspond to one of the estimated \(\beta\)’s in the equation above.
When fitting a regression model in R with two explanatory variables A and B, and their interaction, these three are equivalent:
- y ~ A + B + A:B
- y ~ A + B + A*B
- y ~ A*B
Recall your table of descriptive statistics - map each coefficient from the summary() output from “cog_mdl” to the group means.
Interpret your coefficients in the context of the study.
Recall that we can obtain our parameter estimates using various functions such as summary(),coef(), coefficients(), etc.
You may find it helpful to review your descriptive statistics from Q4, or the findings from your mapping exercise in Q6.
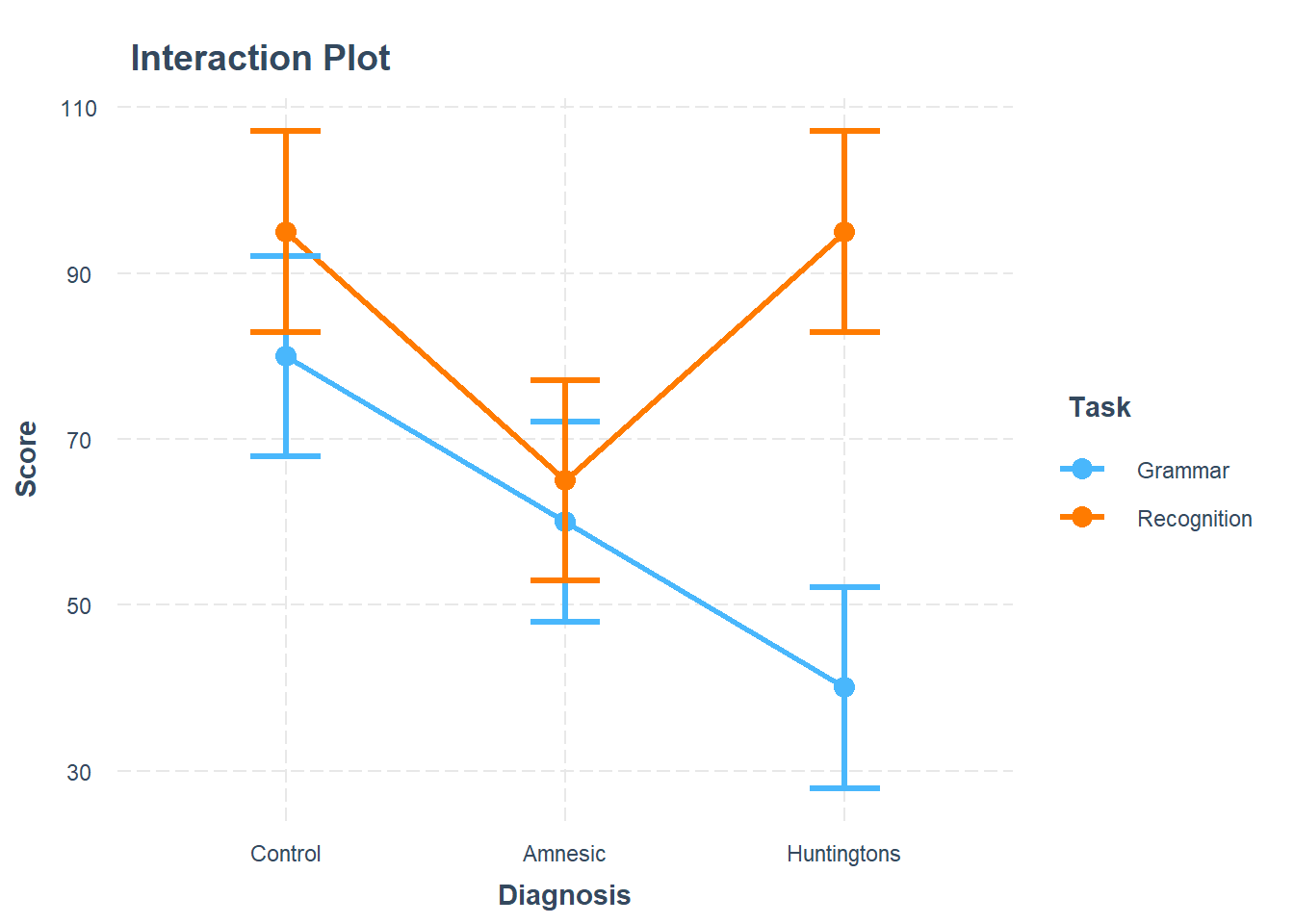
Visualise Interaction Model
Using the cat_plot() function from the interactions package, visualise the interaction effects from your model.
Try to summarise the interaction effects in a few short and concise sentences.
In terms of of specification, it might be useful to look up the helper function. As a quick guide:
-
model= name model to be used -
pred= The categorical predictor variable that will appear on the x-axis -
modx= The categorical moderator variable
Remember to give your plot informative titles/labels. You, for example, likely want to give your plot:
- a clear and concise title (specify
main.title =) - axis labels with units or scale included (specify
x.label =andy.label =) - a legend title (specify
legend.main =)
Writing Up & Presenting Results
Provide key model results in a formatted table.
Use tab_model() from the sjPlot package.
Remember that you can rename your DV and IV labels by specifying dv.labels and pred.labels.
Interpret your results in the context of the research question and report your model in full.
Make reference to the interaction plot and regression table.
Compile Report
Knit your report to PDF, and check over your work. To do so, you should make sure:
- Only the output you want your reader to see is visible (e.g., do you want to hide your code?)
- Check that the tinytex package is installed
- Ensure that the ‘yaml’ (bit at the very top of your document) looks something like this:
---
title: "this is my report title"
author: "B1234506"
date: "07/09/2024"
output: bookdown::pdf_document2
---If you are having issues knitting directly to PDF, try the following:
- Knit to HTML file
- Open your HTML in a web-browser (e.g. Chrome, Firefox)
- Print to PDF (Ctrl+P, then choose to save to PDF)
- Open file to check formatting
To not show the code of an R code chunk, and only show the output, write:
```{r, echo=FALSE}
# code goes here
```To show the code of an R code chunk, but hide the output, write:
```{r, results='hide'}
# code goes here
```To hide both code and output of an R code chunk, write:
```{r, include=FALSE}
# code goes here
```You must make sure you have tinytex installed in R so that you can “Knit” your Rmd document to a PDF file:
install.packages("tinytex")
tinytex::install_tinytex()
Footnotes
Some researchers may point out that a design where each person was assessed on both tasks might have been more efficient. However, the task factor in such design would then be within-subjects, meaning that the scores corresponding to the same person would be correlated. To analyse such design we will need a different method which (spoiler alert!) will be discussed next year in DAPR3.↩︎