| variable | description |
|---|---|
| ape | Ape Name |
| species | Species (Bonobo, Chimpanzee, Gorilla, Orangutan) |
Week 2 Exercises: Logistic and Longitudinal
Great Apes!
Data: msmr_apespecies.csv & msmr_apeage.csv
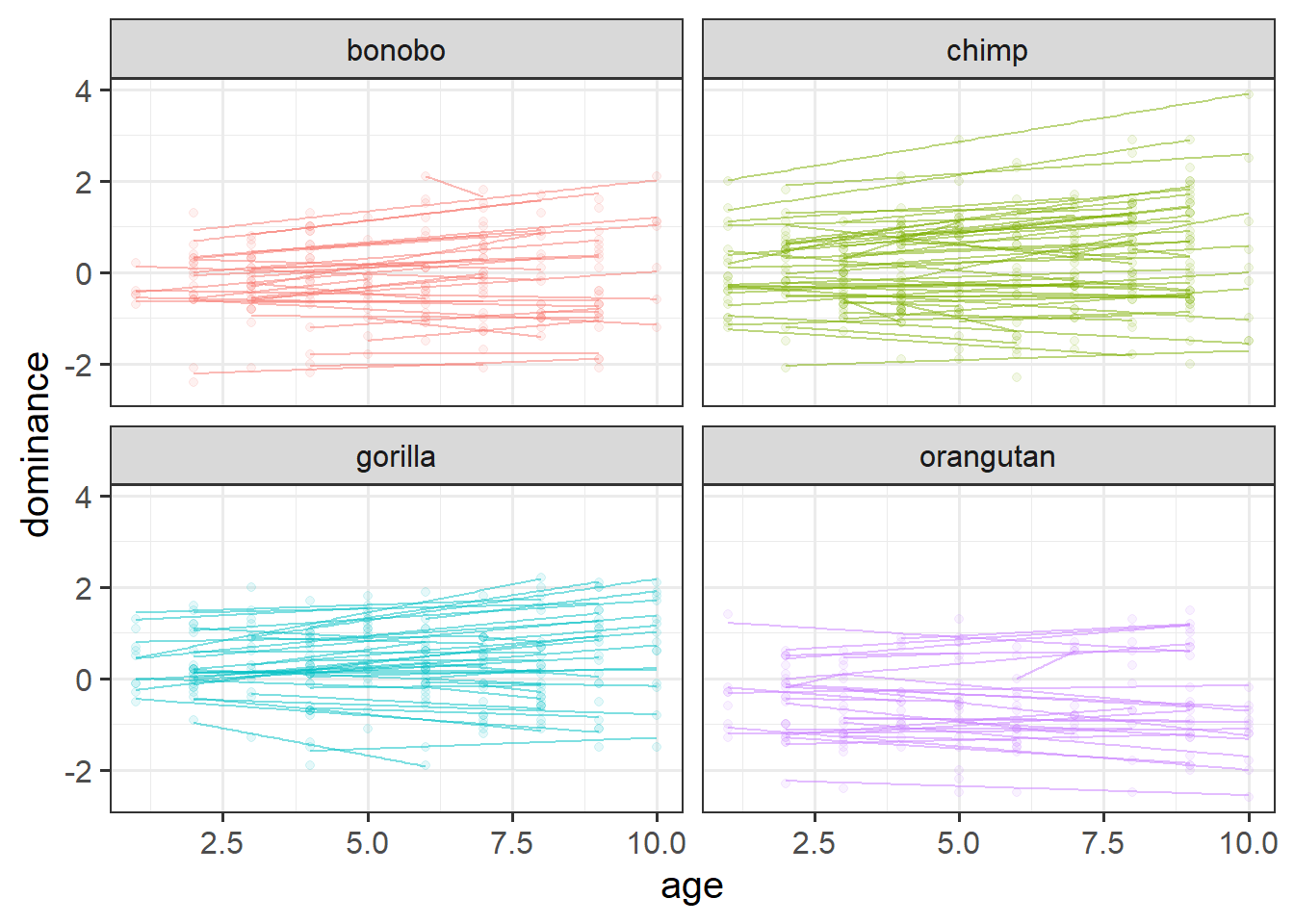
We have data from a large sample of great apes who have been studied between the ages of 1 to 10 years old (i.e. during adolescence). Our data includes 4 species of great apes: Chimpanzees, Bonobos, Gorillas and Orangutans. Each ape has been assessed on a primate dominance scale at various ages. Data collection was not very rigorous, so apes do not have consistent assessment schedules (i.e., one may have been assessed at ages 1, 3 and 6, whereas another at ages 2 and 8).
The researchers are interested in examining how the adolescent development of dominance in great apes differs between species.
Data on the dominance scores of the apes are available at https://uoepsy.github.io/data/msmr_apeage.csv and the information about which species each ape is are in https://uoepsy.github.io/data/msmr_apespecies.csv.
| variable | description |
|---|---|
| ape | Ape Name |
| age | Age at assessment (years) |
| dominance | Dominance (Z-scored) |
Read in the data and check over it. Do any relevant cleaning/wrangling that might be necessary.
How is this data structure “hierarchical” (or “clustered”)? What are our level 1 units, and what are our level 2 units?
For how many apes do we have data? How many of each species?
How many datapoints does each ape have?
We’ve seen this last week too - counting the different levels in our data. See 2B #getting-to-know-my-monkeys for an example (with another monkey example!)
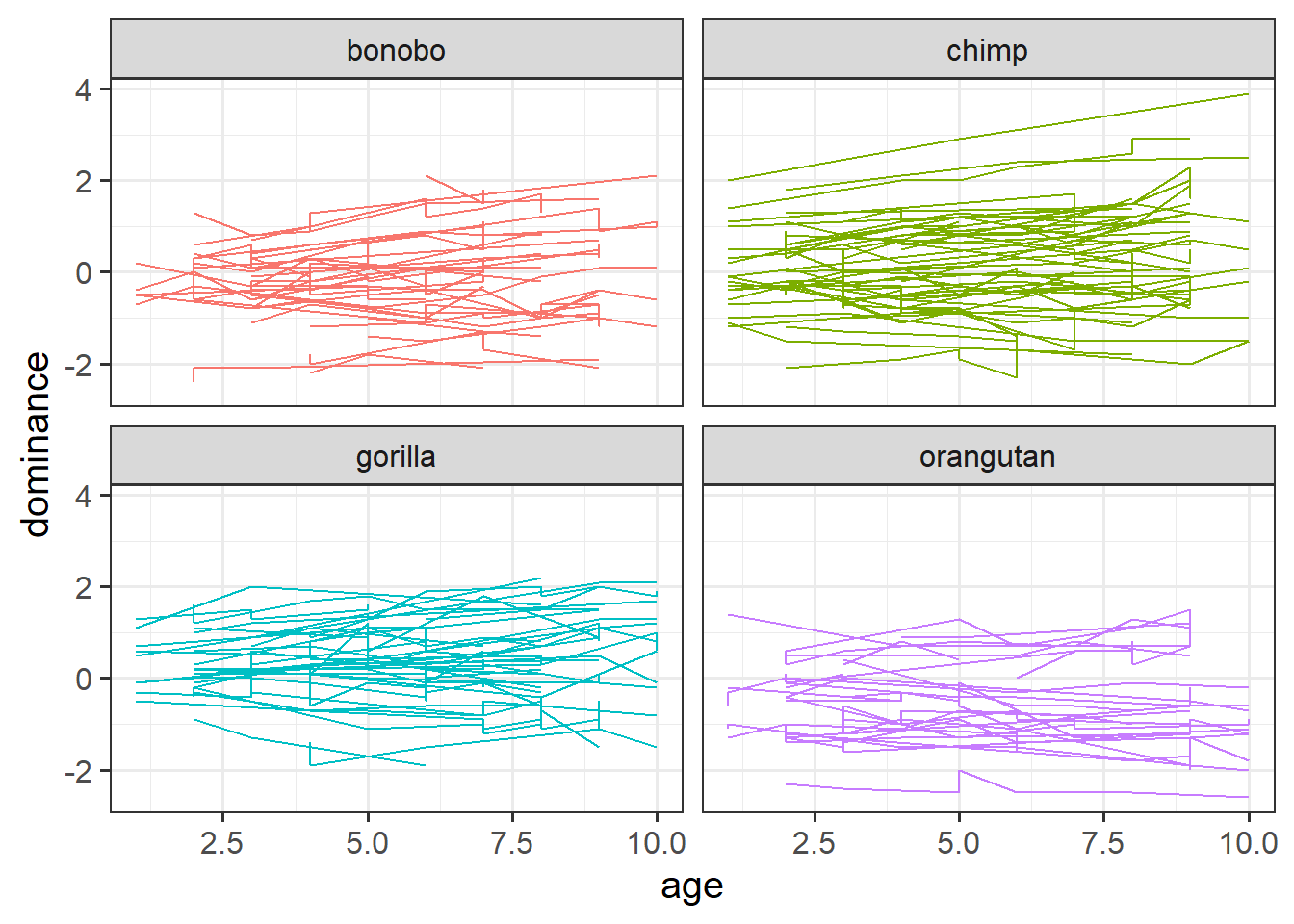
Make a plot to show how dominance changes as apes get older.
In 2B #exploring-the-data we made a facet for each cluster (each participant). That was fine because we had only 20 people. In this dataset we have 168! That’s too many to facet. The group aesthetic will probably help instead!
Recenter the age variable on 1, which is the youngest ages that we’ve got data on for any of our species.
Then fit a model that estimates the differences between primate species in how dominance changes over time.
Do primate species differ in the growth of dominance?
Perform an appropriate test/comparison.
This is asking about the age*species interaction, which in our model is represented by 3 parameters. To assess the overall question, it might make more sense to do a model comparison.
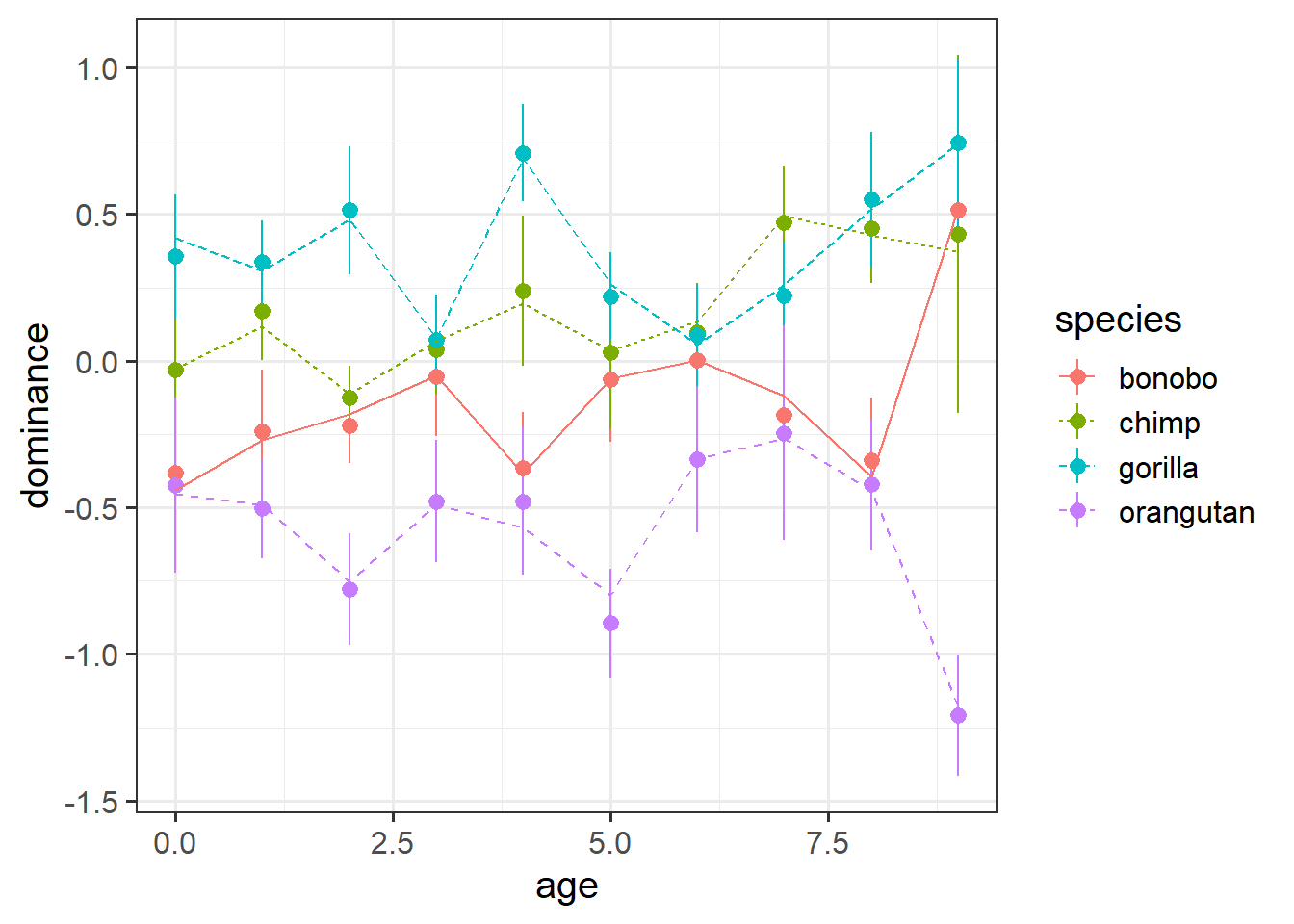
Plot the average model predicted values for each age.
Before you plot.. do you expect to see straight lines? (remember, not every ape is measured at age 2, or age 3, etc).
This is like taking predict() from the model, and then then grouping by age, and calculating the mean of those predictions. However, we can do this more easily using augment() and then some fancy stat_summary() in ggplot (see the lecture).
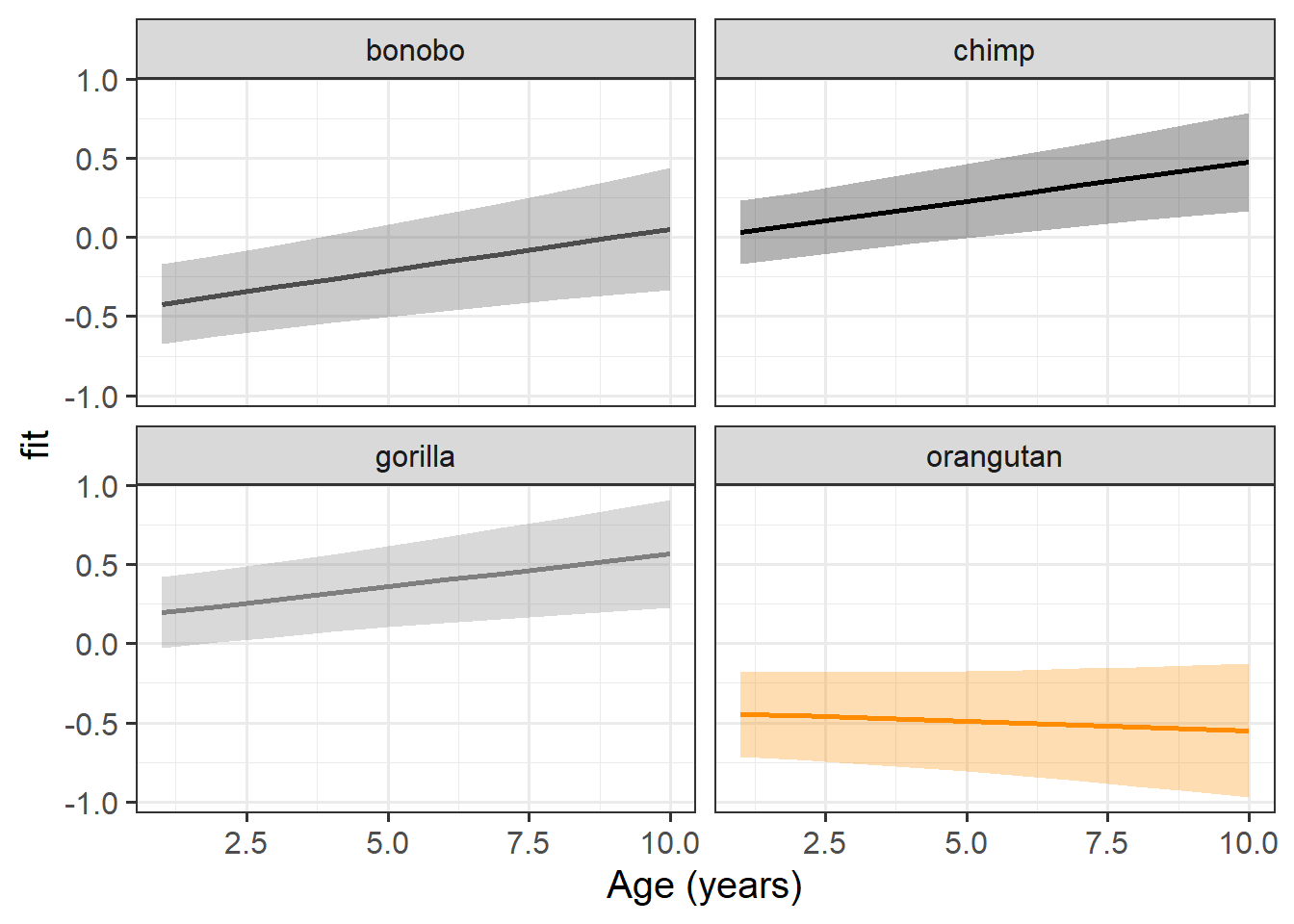
Plot the model based fixed effects:
Interpret each of the fixed effects from the model (you might also want to get some p-values or confidence intervals).
Each of the estimates should correspond to part of our plot from the previous question.
Trolley problems
Data: msmr_trolley.csv
The “Trolley Problem” is a thought experiment in moral philosophy that asks you to decide whether or not to pull a lever to divert a trolley. Pulling the lever changes the trolley direction from hitting 5 people to a track on which it will hit one person.
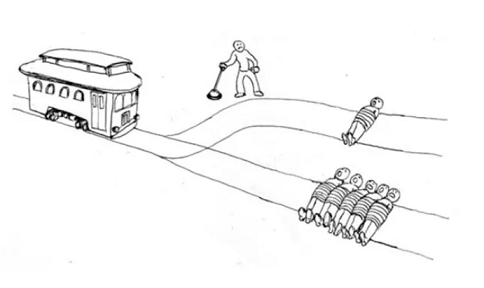
Previous research has found that the “framing” of the problem will influence the decisions people make:
| positive frame | neutral frame | negative frame |
|---|---|---|
| 5 people will be saved if you pull the lever; one person on another track will be saved if you do not pull the lever. All your actions are legal and understandable. Will you pull the lever? | 5 people will be saved if you pull the lever, but another person will die. One people will be saved if you do not pull the lever, but 5 people will die. All your actions are legal and understandable. Will you pull the lever? | One person will die if you pull the lever. 5 people will die if you do not pull the lever. All your actions are legal and understandable. Will you pull the lever? |
We conducted a study to investigate whether the framing effects on moral judgements depends upon the stakes (i.e. the number of lives saved).
120 participants were recruited, and each gave answers to 12 versions of the thought experiment. For each participant, four versions followed each of the positive/neutral/negative framings described above, and for each framing, 2 would save 5 people and 2 would save 15 people.
The data are available at https://uoepsy.github.io/data/msmr_trolley.csv.
| variable | description |
|---|---|
| PID | Participant ID |
| frame | framing of the thought experiment (positive/neutral/negative |
| lives | lives at stake in the thought experiment (5 or 15) |
| lever | Whether or not the participant chose to pull the lever (1 = yes, 0 = no) |
Read in the data and check over how many people we have, and whether we have complete data for each participant.
I would maybe try data |> group_by(participant) |> summarise(), and then use the n_distinct() function to count how many “things” each person sees (e.g., 2B #example).
Construct an appropriate plot to summarise the data in a suitable way to illustrate the research question.
Something making use of stat_summary() to give proportions, a bit like the plot in 2B #getting-to-know-my-monkeys?
Fit a model to assess the research aims.
Don’t worry if it gives you an error, we’ll deal with that in a second.
- Remember, a good way to start is to split this up into 3 parts: 1) the outcome and fixed effects, 2) the grouping structure, and 3) the random slopes.
- fitting (or attempting to fit!)
glmermodels might take time!
This is probably the first time we’ve had to deal with a model not converging.
While sometimes changing the optimizer can help, more often than not, the model we are trying to fit is just too complex. Often, the groups in our sample just don’t vary enough for us to estimate a random slope.
The aim here is to simplify our random effect structure in order to obtain a converging model, but be careful not to over simplify.
Try it now. What model do you end up with? (You might not end up with the same model as each other, which is fine. These methods don’t have “cookbook recipes”!)
you could think of the interaction as the ‘most complex’ part of our random effects, so you might want to remove that first.
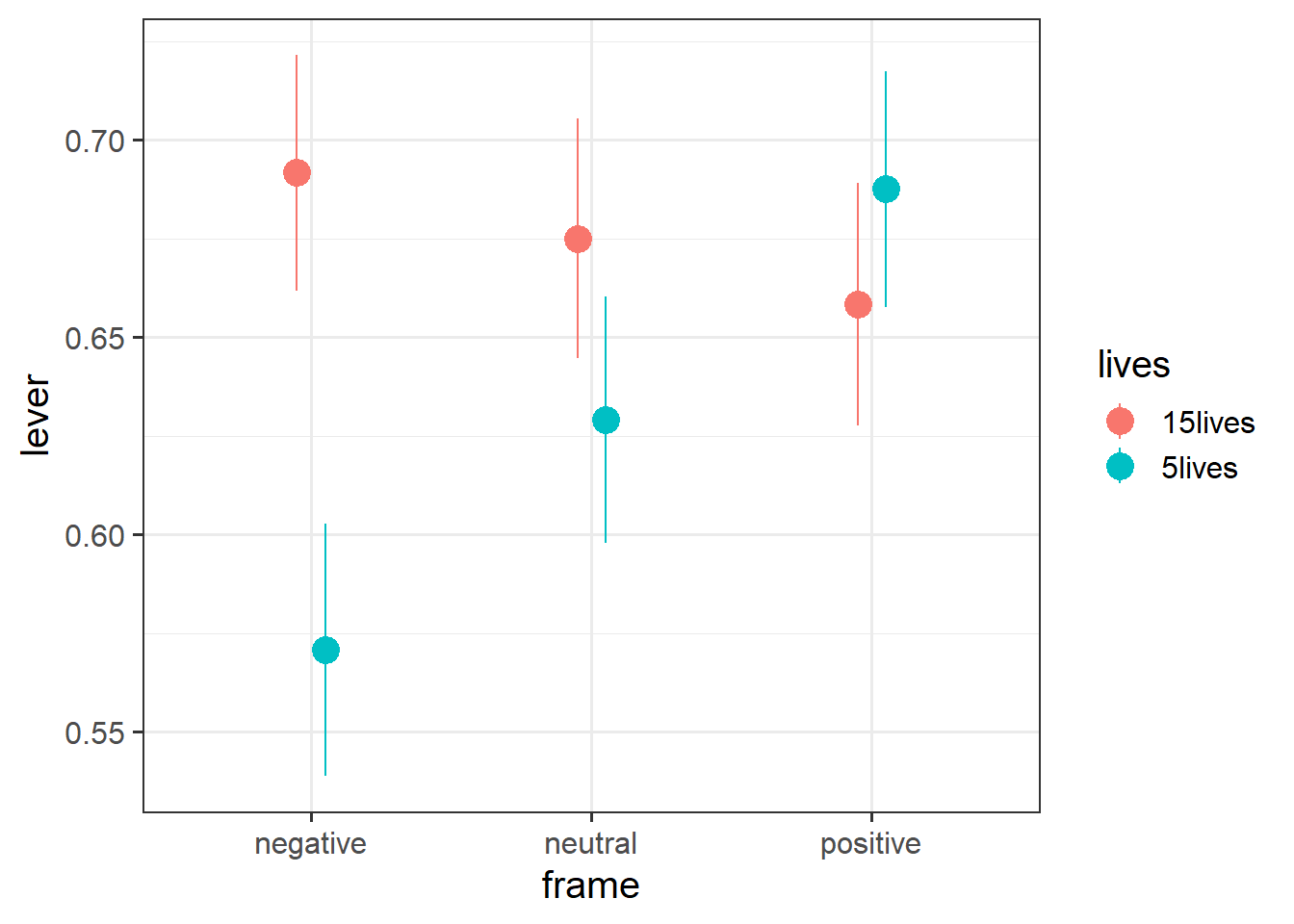
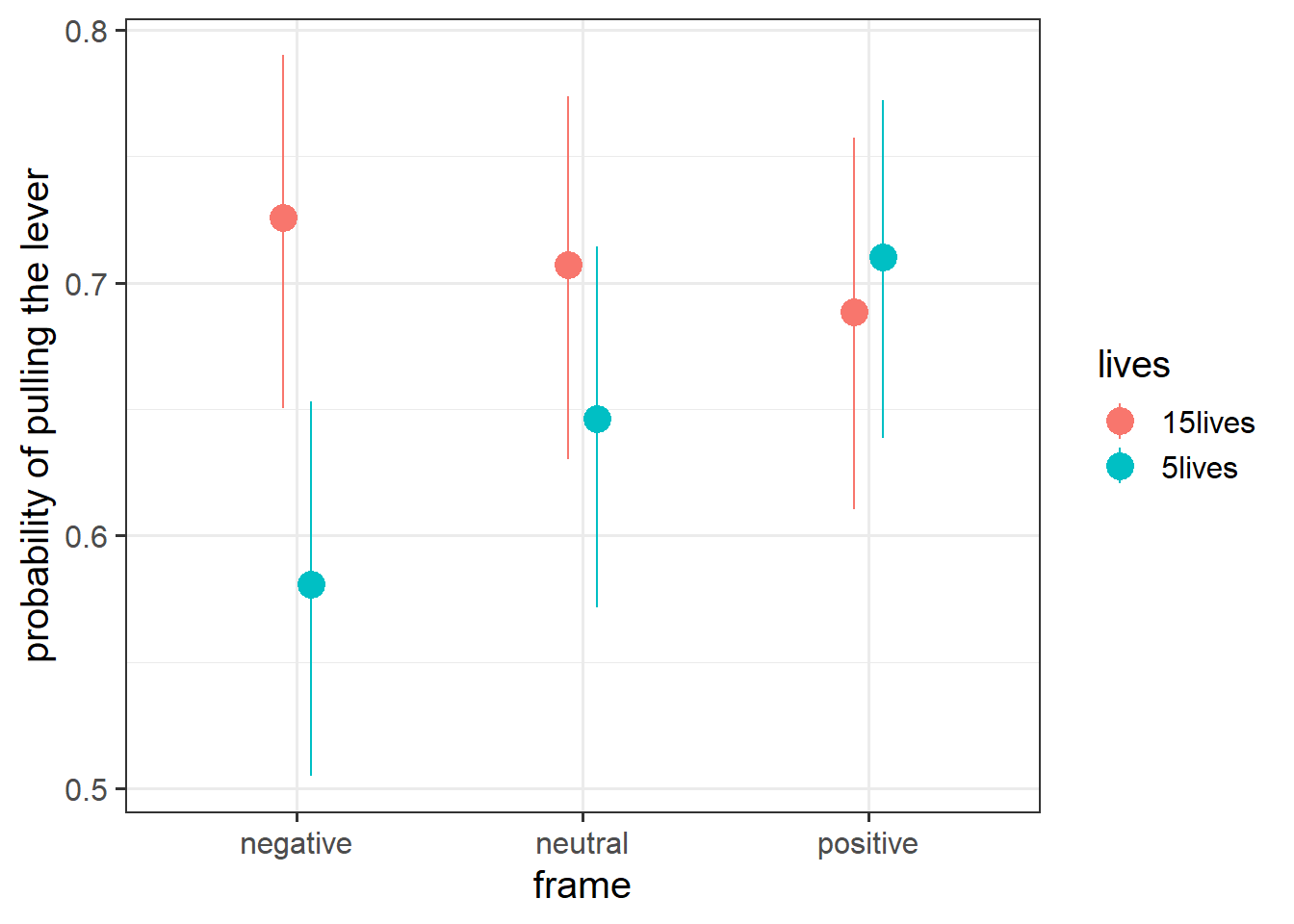
Plot the predicted probabilities from your model for each combination of frame and lives.
Optional extra: Novel Word Learning
Data: nwl.Rdata
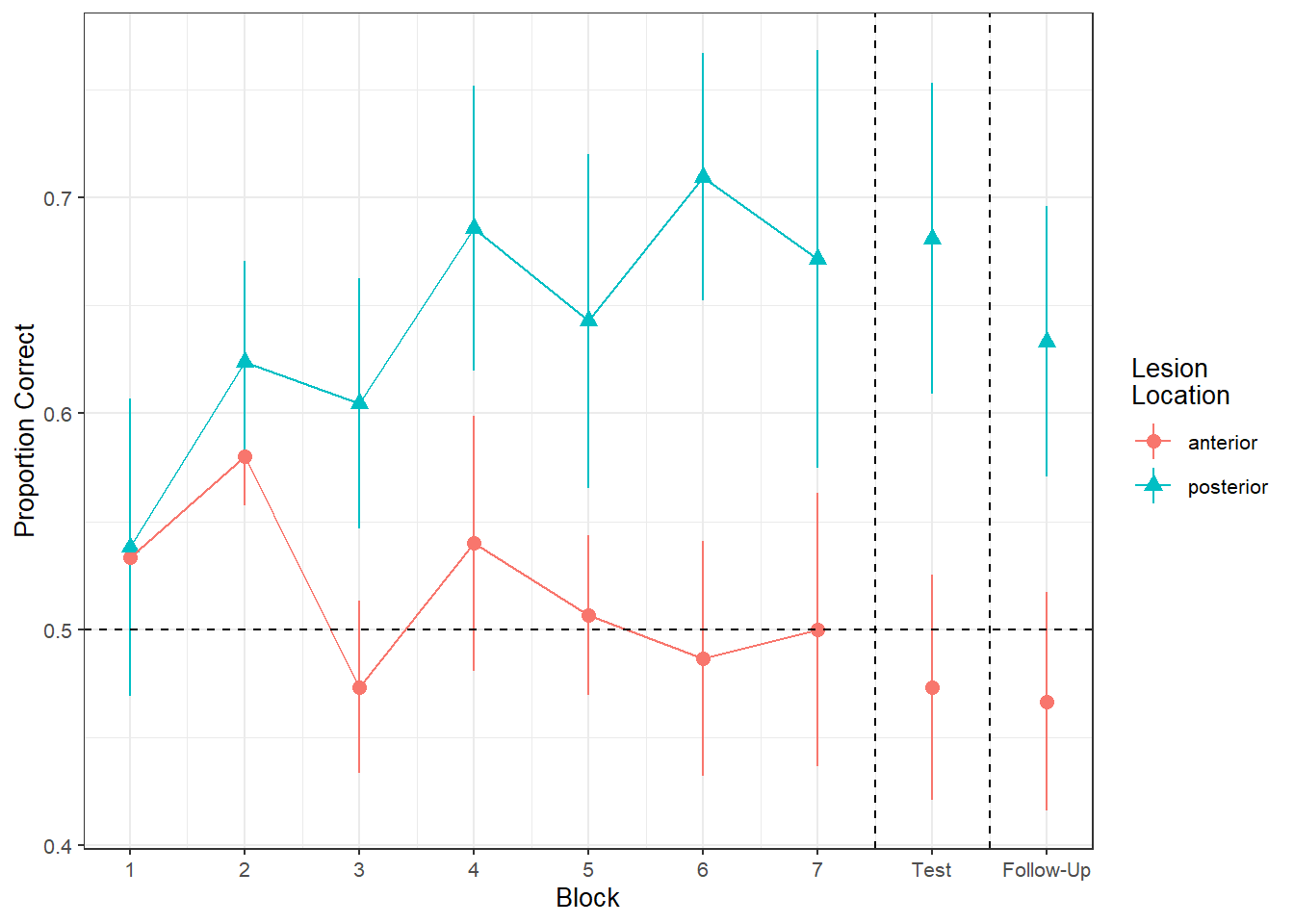
load(url("https://uoepsy.github.io/msmr/data/nwl.RData"))In the nwl data set (accessed using the code above), participants with aphasia are separated into two groups based on the general location of their brain lesion: anterior vs. posterior. There is data on the numbers of correct and incorrect responses participants gave in each of a series of experimental blocks. There were 7 learning blocks, immediately followed by a test. Finally, participants also completed a follow-up test.
Data were also collect from healthy controls.
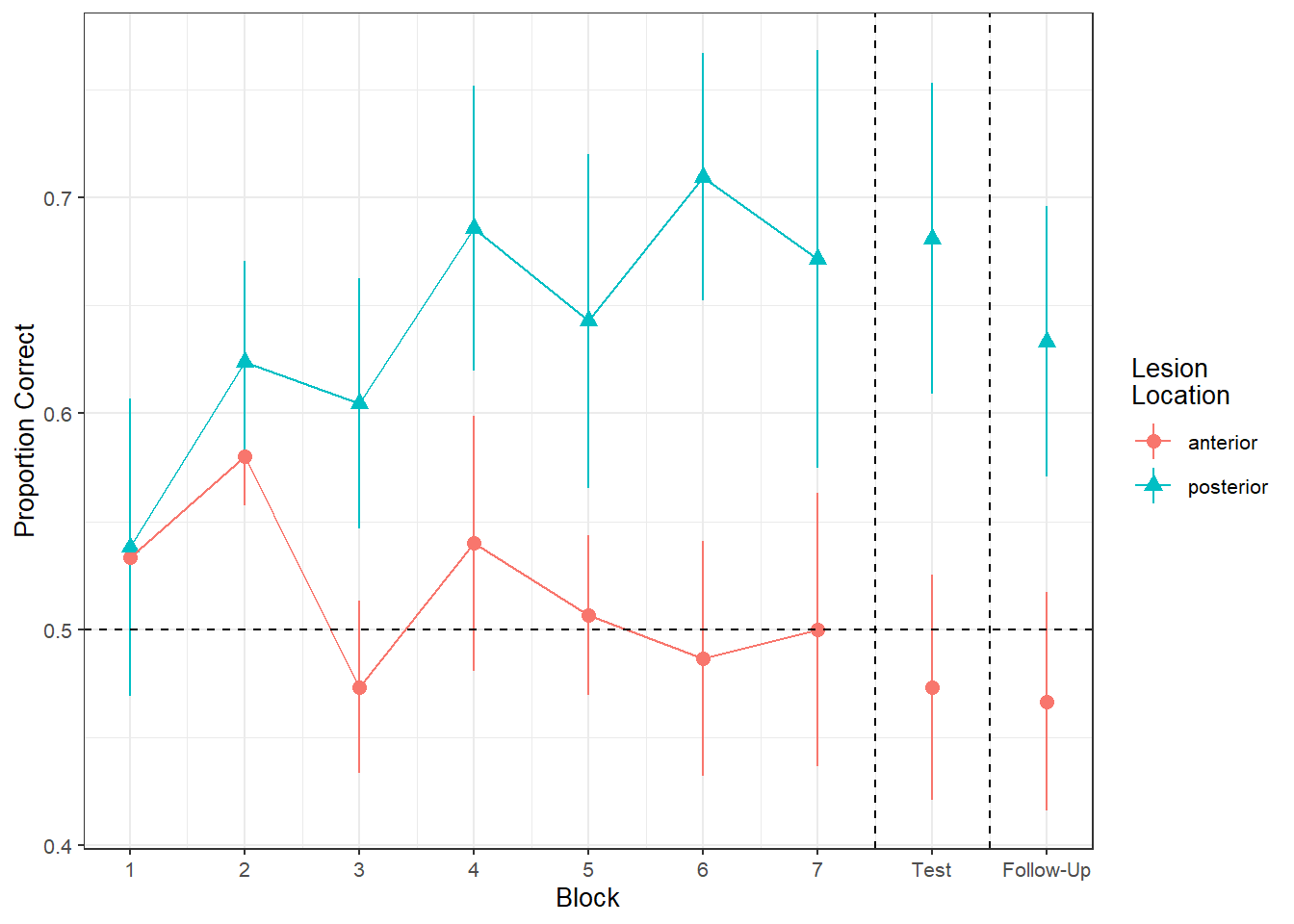
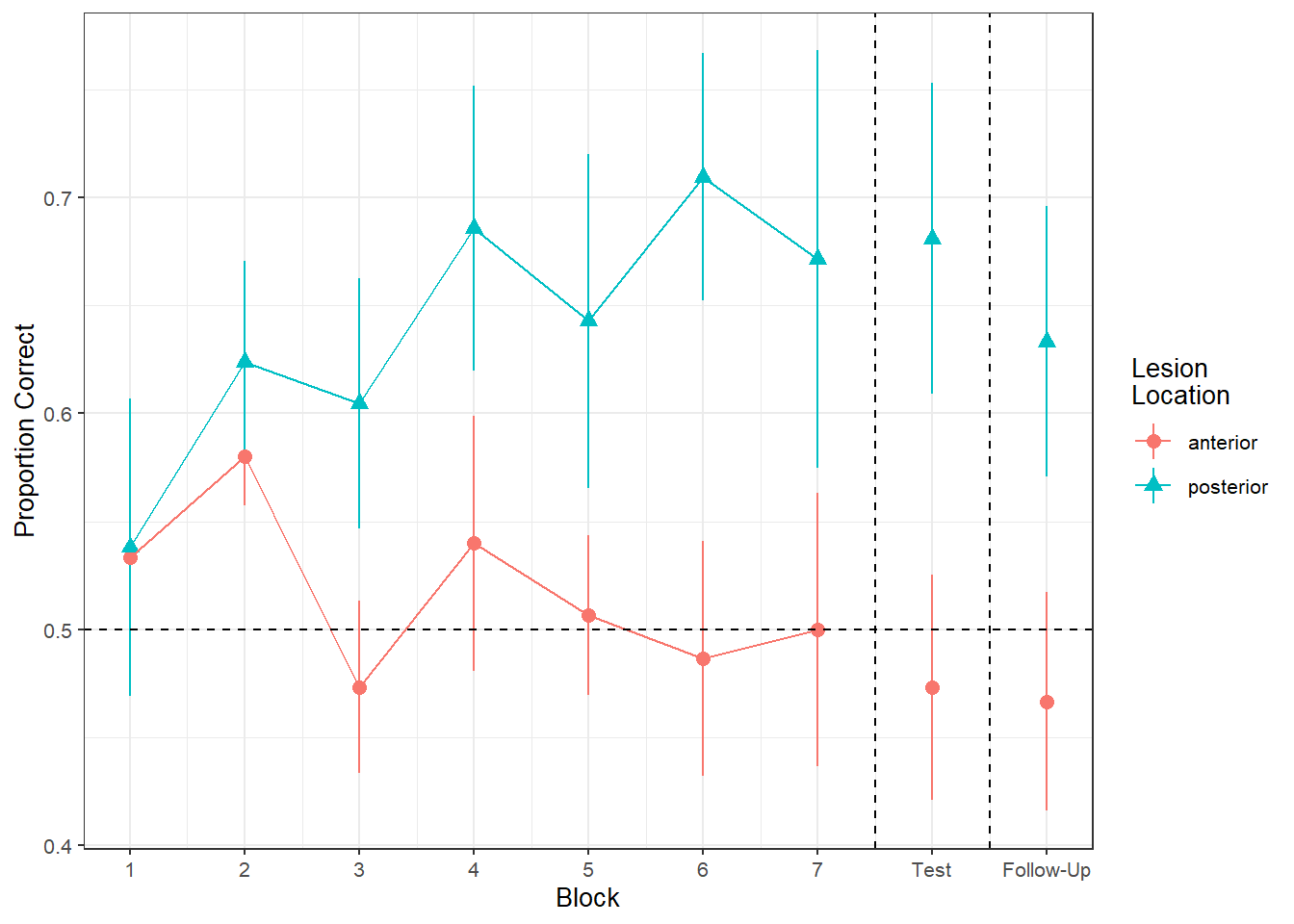
Figure 1 shows the differences between lesion location groups in the average proportion of correct responses at each point in time (i.e., each block, test, and follow-up)
| variable | description |
|---|---|
| group | Whether participant is a stroke patient ('patient') or a healthy control ('control') |
| lesion_location | Location of brain lesion: anterior vs posterior |
| block | Experimental block (1-9). Blocks 1-7 were learning blocks, immediately followed by a test in block 8. Block 9 was a follow-up test at a later point |
| PropCorrect | Proportion of 30 responses in a given block that the participant got correct |
| NumCorrect | Number of responses (out of 30) in a given block that the participant got correct |
| NumError | Number of responses (out of 30) in a given block that the participant got incorrect |
| ID | Participant Identifier |
| Phase | Experimental phase, corresponding to experimental block(s): 'Learning', 'Immediate','Follow-up' |
Load the data. Take a look around. Any missing values? Can you think of why?
Our broader research aim today is to compare the two lesion location groups (those with anterior vs. posterior lesions) with respect to their accuracy of responses over the course of the study.
- What is the outcome variable?
Think carefully: there might be several variables which either fully or partly express the information we are considering the “outcome” here. We saw this back in USMR with the glm()!
Research Question 1:
Is the learning rate (training blocks) different between the two lesion location groups?
Do we want
cbind(num_successes, num_failures)?Ensure you are running models on only the data we are actually interested in.
- Are the healthy controls included in the research question under investigation?
- Are the testing blocks included in the research question, or only the learning blocks?
We could use model comparison via likelihood ratio tests (using
anova(model1, model2, model3, ...). For this question, we could compare:- A model with just the change over the sequence of blocks
- A model with the change over the sequence of blocks and an overall difference between groups
- A model with groups differing with respect to their change over the sequence of blocks
What about the random effects part?
- What are our observations grouped by?
- What variables can vary within these groups?
- What do you want your model to allow to vary within these groups?
Research Question 2
In the testing phase, does performance on the immediate test differ between lesion location groups, and does the retention from immediate to follow-up test differ between the two lesion location groups?
Let’s try a different approach to this. Instead of fitting various models and comparing them via likelihood ratio tests, just fit the one model which could answer both parts of the question above.
- This might required a bit more data-wrangling beforehand. Think about the order of your factor levels (alphabetically speaking, “Follow-up” comes before “Immediate”)!
- In
family = binomial(link='logit'). What function is used to relate the linear predictors in the model to the expected value of the response variable?
- How do we convert this into something more interpretable?
Make sure you pay attention to trying to interpret each fixed effect from your models.
These can be difficult, especially when it’s logistic, and especially when there are interactions.
- What is the increase in the odds of answering correctly in the immediate test if you were to have a posterior legion instead of an anterior legion?
Recreate the visualisation in Figure 2.