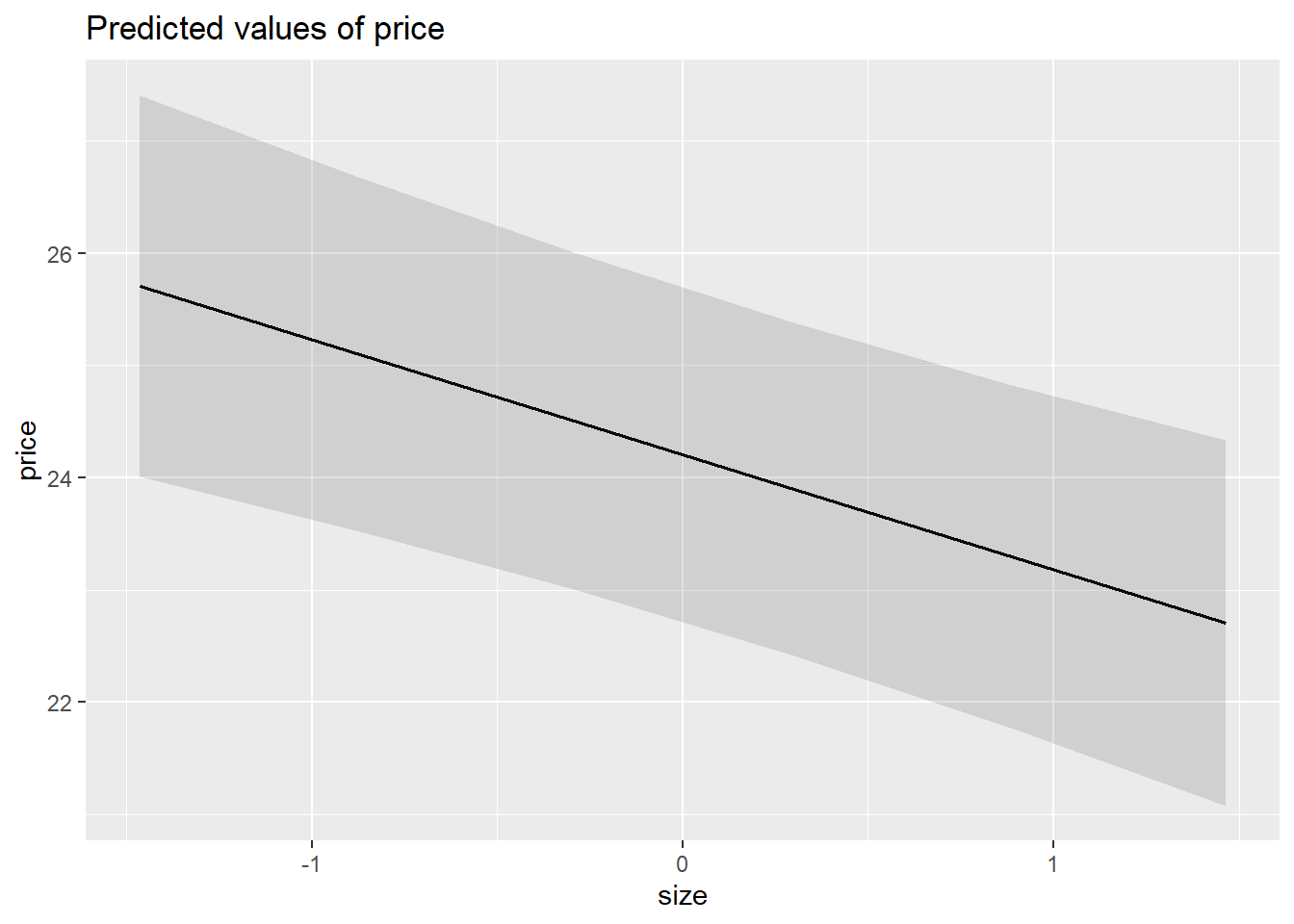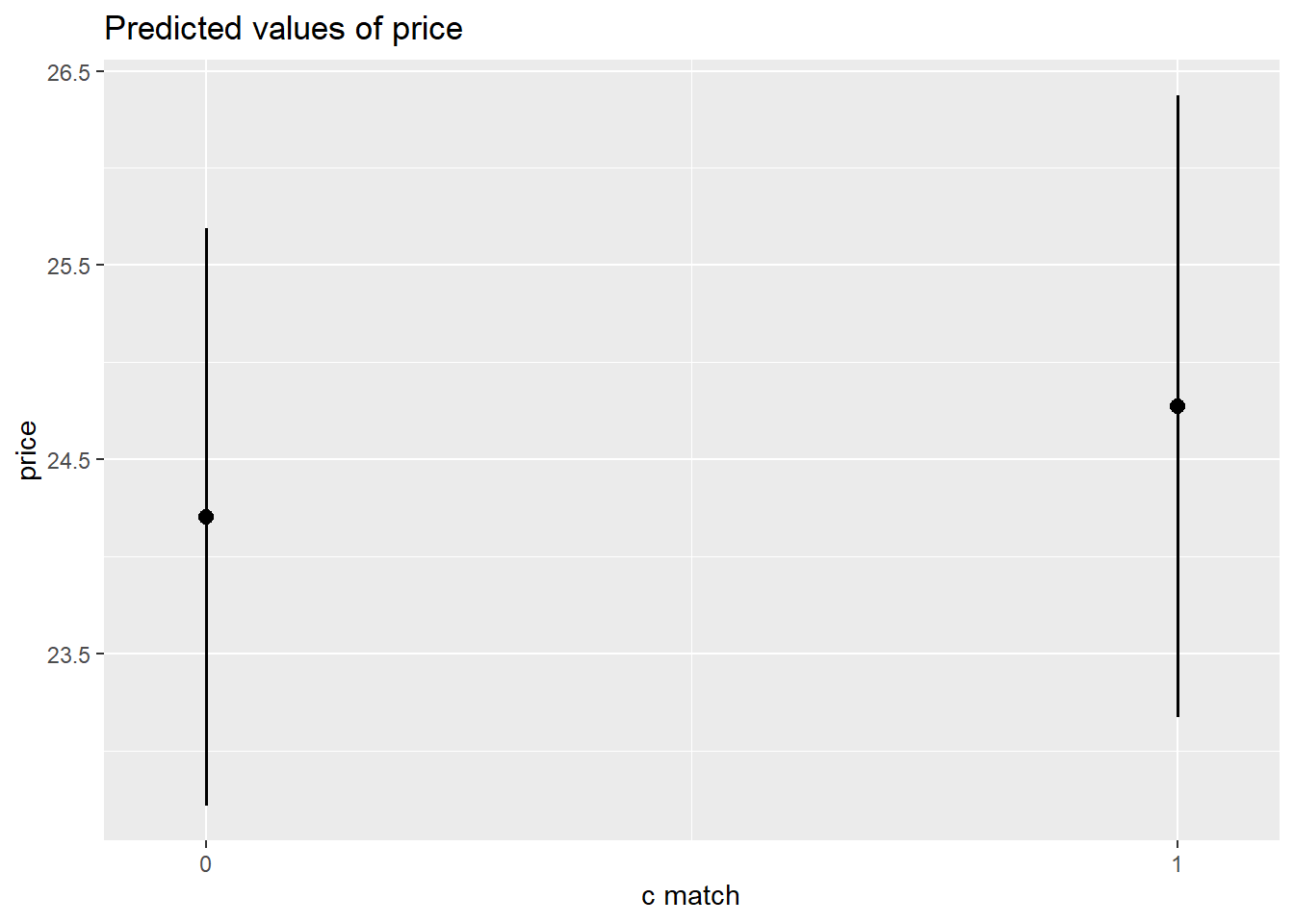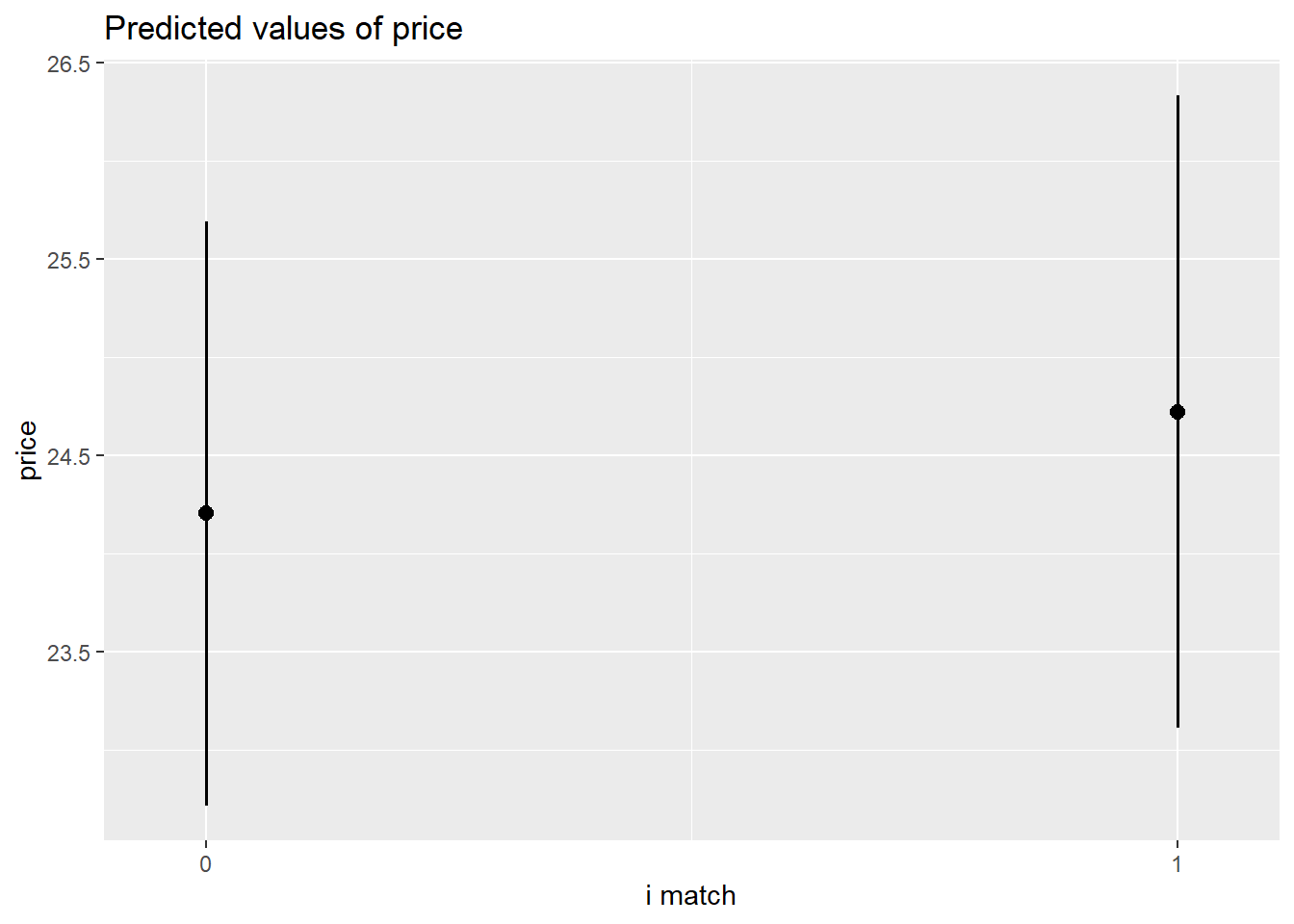Each of these pages provides an analysis run through for a different type of design. Each document is structured in the same way:
- First the data and research context is introduced. For the purpose of these tutorials, we will only use examples where the data can be shared - either because it is from an open access publication, or because it is unpublished or simulated.
- Second, we go through any tidying of the data that is required, before creating some brief descriptives and visualizations of the raw data.
- Then, we conduct an analysis. Where possible, we translate the research questions into formal equations prior to fitting the models in lme4. Model comparisons are conducted, along with checks of distributional assumptions on our model residuals.
- Finally, we visualize and interpret our analysis.
Please note that there will be only minimal explanation of the steps undertaken here, as these pages are intended as example analyses rather than additional labs readings. Please also be aware that there are many decisions to be made throughout conducting analyses, and it may be the case that you disagree with some of the choices we make here. As always with these things, it is how we justify our choices that is important. We warmly welcome any feedback and suggestions to improve these examples: please email ug.ppls.stats@ed.ac.uk.
Overview
This data comes from an undergraduate dissertation student. She ran an experiment looking at the way people’s perception of the size of models influences the price they are willing to pay for products. Participants saw a series of pictures of a number of items of clothing. The images had been manipulated so that (a) all pictures were of the same model, and (b) the size of the model differed from a 6 to 16. In total participants saw 54 images. During the study, each picture was presented to participants with a sliding scale from 0 to 100 underneath. Participants simply had to drag the cursor to the appropriate point on the slide to indicate how much they would pay for the garment.
This data was collected using Qualtrics. The resultant output was a wide format data set where each participant (n=120) is a row and each item (n=54) is a column. Along with each of these questions were a series of demographic variables, and two short survey measures assessing participants attitude to thinness.
The main ideas we were interested in looking at were whether:
- participants would pay more for garments on thinner models
- participants would pay more for items when the model size matched their actual size
- participants would pay more for items when the model size matched their ideal size
- the extent to which participants idealize thin figures would moderate (ii)
Data Wrangling
Unlike some of our examples, there was some fairly serious data cleaning needed with this data set. So let’s work through it.
The data is available at https://uoepsy.github.io/data/data_HA.csv
Code
library(tidyverse)
df <- read_csv("https://uoepsy.github.io/data/data_HA.csv")
head(df)
# A tibble: 6 × 116
Respons…¹ Gender Age Dress…² Top_1…³ Botto…⁴ Dress…⁵ Botto…⁶ Top_6…⁷ Dress…⁸
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 P_ID1001 1 22 45 24 30 30 20 17 15
2 P_ID1002 1 20 30 19 9 52 0 5 16
3 P_ID1003 1 21 30 12 4 18 20 8 12
4 P_ID1004 1 21 39 30 30 45 25 20 25
5 P_ID1005 1 23 10 5 15 7 15 5 20
6 P_ID1006 1 43 40 30 45 60 35 25 30
# … with 106 more variables: Dress_12...11 <dbl>, Bottom_12...12 <dbl>,
# Top_10...13 <dbl>, Top_8...14 <dbl>, Bottom_6...15 <dbl>,
# Bottom_8...16 <dbl>, Dress_14...17 <dbl>, Top_16...18 <dbl>,
# Bottom_6...19 <dbl>, Bottom_14...20 <dbl>, Top_8...21 <dbl>,
# Dress_8...22 <dbl>, Top_12...23 <dbl>, Top_14...24 <dbl>,
# Dress_16...25 <dbl>, Top_10...26 <dbl>, Top_14...27 <dbl>,
# Dress_8...28 <dbl>, Dress_16...29 <dbl>, Bottom_12...30 <dbl>, …
First things first, let’s use a handy package to change all our variable names to a nice easy “snake_case”:
Code
library(janitor)
df <- clean_names(df)
head(df)
# A tibble: 6 × 116
respons…¹ gender age dress…² top_1…³ botto…⁴ dress…⁵ botto…⁶ top_6_9 dress…⁷
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 P_ID1001 1 22 45 24 30 30 20 17 15
2 P_ID1002 1 20 30 19 9 52 0 5 16
3 P_ID1003 1 21 30 12 4 18 20 8 12
4 P_ID1004 1 21 39 30 30 45 25 20 25
5 P_ID1005 1 23 10 5 15 7 15 5 20
6 P_ID1006 1 43 40 30 45 60 35 25 30
# … with 106 more variables: dress_12_11 <dbl>, bottom_12_12 <dbl>,
# top_10_13 <dbl>, top_8_14 <dbl>, bottom_6_15 <dbl>, bottom_8_16 <dbl>,
# dress_14_17 <dbl>, top_16_18 <dbl>, bottom_6_19 <dbl>, bottom_14_20 <dbl>,
# top_8_21 <dbl>, dress_8_22 <dbl>, top_12_23 <dbl>, top_14_24 <dbl>,
# dress_16_25 <dbl>, top_10_26 <dbl>, top_14_27 <dbl>, dress_8_28 <dbl>,
# dress_16_29 <dbl>, bottom_12_30 <dbl>, dress_6_31 <dbl>,
# bottom_14_32 <dbl>, bottom_8_33 <dbl>, top_16_34 <dbl>, …
Our next job is to cut out a few variables that we will not work with in this example. These are a set of questions asking about clothing preferences. They all end in p and appear at the end of the data set.
Code
df1 <-
df %>%
select(-ends_with("p"))
Now we want to create scores for the two surveys scored using Likert-type scales. We’re going to take the means of these scales as our scores:
Code
# TI score
df1 <-
df1 %>%
select(contains("_tii")) %>%
rowMeans(., na.rm=T) %>%
bind_cols(df1, tii_score = .)
# MA score
df1 <-
df1 %>%
select(contains("_mo")) %>%
rowMeans(., na.rm=T) %>%
bind_cols(df1, ma_score = .)
Next, we need to make some changes to the coding of current and ideal size
Code
df1 <-
df1 %>%
mutate(
c.size = recode(current_size, 6, 8, 10, 12, 14, 16),
i.size = recode(ideal_size, 6, 8, 10, 12, 14, 16)
)
Always sensible to check our changes:
Code
df1 %>%
select(current_size, ideal_size, c.size, i.size)
# A tibble: 120 × 4
current_size ideal_size c.size i.size
<dbl> <dbl> <dbl> <dbl>
1 3 2 10 8
2 1 1 6 6
3 2 1 8 6
4 2 1 8 6
5 3 2 10 8
6 4 3 12 10
7 3 2 10 8
8 2 2 8 8
9 5 3 14 10
10 3 2 10 8
# … with 110 more rows
Excellent, that has worked.
Our next step (and this one is not strictly necessary) is that we are going to aggregate over the different types of top, bottoms and dresses. We could treat each individual garment of clothing as exactly that, but in this instance it was decided that this was not of interest, and to simplify the models, we would create scores for each broad category by size.
Code
df1 <-
df1 %>%
mutate(top_S6 = rowMeans(.[names(df1)[grepl("top_6",names(df1))]]),
top_S8 = rowMeans(.[names(df1)[grepl("top_8",names(df1))]]),
top_S10 = rowMeans(.[names(df1)[grepl("top_10",names(df1))]]),
top_S12 = rowMeans(.[names(df1)[grepl("top_12",names(df1))]]),
top_S14 = rowMeans(.[names(df1)[grepl("top_14",names(df1))]]),
top_S16 = rowMeans(.[names(df1)[grepl("top_16",names(df1))]]),
bottom_S6 = rowMeans(.[names(df1)[grepl("bottom_6",names(df1))]]),
bottom_S8 = rowMeans(.[names(df1)[grepl("bottom_8",names(df1))]]),
bottom_S10 = rowMeans(.[names(df1)[grepl("bottom_10",names(df1))]]),
bottom_S12 = rowMeans(.[names(df1)[grepl("bottom_12",names(df1))]]),
bottom_S14 = rowMeans(.[names(df1)[grepl("bottom_14",names(df1))]]),
bottom_S16 = rowMeans(.[names(df1)[grepl("bottom_16",names(df1))]]),
dress_S6 = rowMeans(.[names(df1)[grepl("dress_6",names(df1))]]),
dress_S8 = rowMeans(.[names(df1)[grepl("dress_8",names(df1))]]),
dress_S10 = rowMeans(.[names(df1)[grepl("dress_10",names(df1))]]),
dress_S12 = rowMeans(.[names(df1)[grepl("dress_12",names(df1))]]),
dress_S14 = rowMeans(.[names(df1)[grepl("dress_14",names(df1))]]),
dress_S16 = rowMeans(.[names(df1)[grepl("dress_16",names(df1))]]),
)
At this point, our next big step is to make the data long. But I am also going to make this a little more manageable by trimming out the variables we do not need. We want ID and age, gender is constant (all female sample), so we do not need this, and then we want the variables we have just created.
Code
df2 <-
df1 %>%
select(response_id, age, 99:120)
df2
# A tibble: 120 × 24
response_id age tii_s…¹ ma_sc…² c.size i.size top_S6 top_S8 top_S10 top_S12
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 P_ID1001 22 2.63 1.78 10 8 20.7 16.3 21.7 20
2 P_ID1002 20 3 2.44 6 6 7.67 12.3 10 17.3
3 P_ID1003 21 2.6 1.78 8 6 16.7 10.3 9 11.7
4 P_ID1004 21 2.27 1.78 8 6 23.3 20 18.3 23.3
5 P_ID1005 23 2.87 2.56 10 8 6.33 10.7 11 8.33
6 P_ID1006 43 2.73 2.22 12 10 28.3 25 41.7 30
7 P_ID1007 55 2.93 1.89 10 8 23.3 28.3 23.3 23.3
8 P_ID1008 20 3.03 2.11 8 8 21.3 20.7 19 23
9 P_ID1009 48 2.7 2.89 14 10 45.7 20.3 26.7 57
10 P_ID1010 21 2.37 2 10 8 15.3 16.7 18.3 13.3
# … with 110 more rows, 14 more variables: top_S14 <dbl>, top_S16 <dbl>,
# bottom_S6 <dbl>, bottom_S8 <dbl>, bottom_S10 <dbl>, bottom_S12 <dbl>,
# bottom_S14 <dbl>, bottom_S16 <dbl>, dress_S6 <dbl>, dress_S8 <dbl>,
# dress_S10 <dbl>, dress_S12 <dbl>, dress_S14 <dbl>, dress_S16 <dbl>, and
# abbreviated variable names ¹tii_score, ²ma_score
OK, now we need to get the data from wide to long format.
Code
df_long <-
df2 %>%
pivot_longer(top_S6:dress_S16, names_to = "garment",values_to = "price")
df_long
# A tibble: 2,160 × 8
response_id age tii_score ma_score c.size i.size garment price
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 P_ID1001 22 2.63 1.78 10 8 top_S6 20.7
2 P_ID1001 22 2.63 1.78 10 8 top_S8 16.3
3 P_ID1001 22 2.63 1.78 10 8 top_S10 21.7
4 P_ID1001 22 2.63 1.78 10 8 top_S12 20
5 P_ID1001 22 2.63 1.78 10 8 top_S14 15.3
6 P_ID1001 22 2.63 1.78 10 8 top_S16 17
7 P_ID1001 22 2.63 1.78 10 8 bottom_S6 21.7
8 P_ID1001 22 2.63 1.78 10 8 bottom_S8 18
9 P_ID1001 22 2.63 1.78 10 8 bottom_S10 30
10 P_ID1001 22 2.63 1.78 10 8 bottom_S12 20
# … with 2,150 more rows
This looks good, but we now need to have variables that code for size and item type.
Code
df_analysis <-
df_long %>%
separate(garment, c("item", "size"), "_S")
And check….
Code
# A tibble: 2,160 × 9
response_id age tii_score ma_score c.size i.size item size price
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl>
1 P_ID1001 22 2.63 1.78 10 8 top 6 20.7
2 P_ID1001 22 2.63 1.78 10 8 top 8 16.3
3 P_ID1001 22 2.63 1.78 10 8 top 10 21.7
4 P_ID1001 22 2.63 1.78 10 8 top 12 20
5 P_ID1001 22 2.63 1.78 10 8 top 14 15.3
6 P_ID1001 22 2.63 1.78 10 8 top 16 17
7 P_ID1001 22 2.63 1.78 10 8 bottom 6 21.7
8 P_ID1001 22 2.63 1.78 10 8 bottom 8 18
9 P_ID1001 22 2.63 1.78 10 8 bottom 10 30
10 P_ID1001 22 2.63 1.78 10 8 bottom 12 20
# … with 2,150 more rows
Finally, our last couple of steps. First, we want to calculate a couple of binary variables that code whether a give item being rated matches a participants actual or ideal size; second, we need to make our item variable a factor, and third, let’s standardize our scale scores for idealization of thinness.
Code
df_analysis <-
df_analysis %>%
mutate(
c.match = factor(if_else(c.size == size, 1, 0)),
i.match = factor(if_else(i.size == size, 1, 0)),
sizefactor = factor(size, levels=c("6","8","10","12","14","16")),
size = scale(as.numeric(as.character(size))),
item = as.factor(item),
id = response_id,
tii_scorez = scale(tii_score),
ma_scorez = scale(ma_score)
)
df_analysis[,c(1,5:11)]
# A tibble: 2,160 × 8
response_id c.size i.size item size[,1] price c.match i.match
<chr> <dbl> <dbl> <fct> <dbl> <dbl> <fct> <fct>
1 P_ID1001 10 8 top -1.46 20.7 0 0
2 P_ID1001 10 8 top -0.878 16.3 0 1
3 P_ID1001 10 8 top -0.293 21.7 1 0
4 P_ID1001 10 8 top 0.293 20 0 0
5 P_ID1001 10 8 top 0.878 15.3 0 0
6 P_ID1001 10 8 top 1.46 17 0 0
7 P_ID1001 10 8 bottom -1.46 21.7 0 0
8 P_ID1001 10 8 bottom -0.878 18 0 1
9 P_ID1001 10 8 bottom -0.293 30 1 0
10 P_ID1001 10 8 bottom 0.293 20 0 0
# … with 2,150 more rows
There’s a debate to be had about whether we should consider our size variable to be numeric or a factor. It could be argued that the sizes of clothing are distinct categories, and that the scale might not be regular (e.g. size 10 is not the same amount bigger than size 8 as size 8 is from size 6). On the other hand, clothing sizes can also be considered points on an underlying latent continuous variable, which might lead you to want to treat it as numeric. Personally, I would want to know exactly how the participants were presented with the sizes - if they were shown the images and the size of the garment was made explicit for each item, then I would be inclined to consider them distinct categories. However, if participants were not explicitly aware of the size of each item, and it was only the size of the model they saw that varied in their size, I am tempted to treat it numerically.
Descriptives
Let’s look at the average price paid by item type and size:
Code
df_analysis %>%
group_by(size, item) %>%
summarize(
price = mean(price, na.rm=T)
) %>%
arrange(item)
# A tibble: 18 × 3
# Groups: size [6]
size[,1] item price
<dbl> <fct> <dbl>
1 -1.46 bottom 26.1
2 -0.878 bottom 25.7
3 -0.293 bottom 23.7
4 0.293 bottom 24.0
5 0.878 bottom 24.1
6 1.46 bottom 22.5
7 -1.46 dress 28.2
8 -0.878 dress 27.3
9 -0.293 dress 26.2
10 0.293 dress 27.3
11 0.878 dress 25.8
12 1.46 dress 26.5
13 -1.46 top 18.6
14 -0.878 top 18.1
15 -0.293 top 20.0
16 0.293 top 19.1
17 0.878 top 19.2
18 1.46 top 19.3
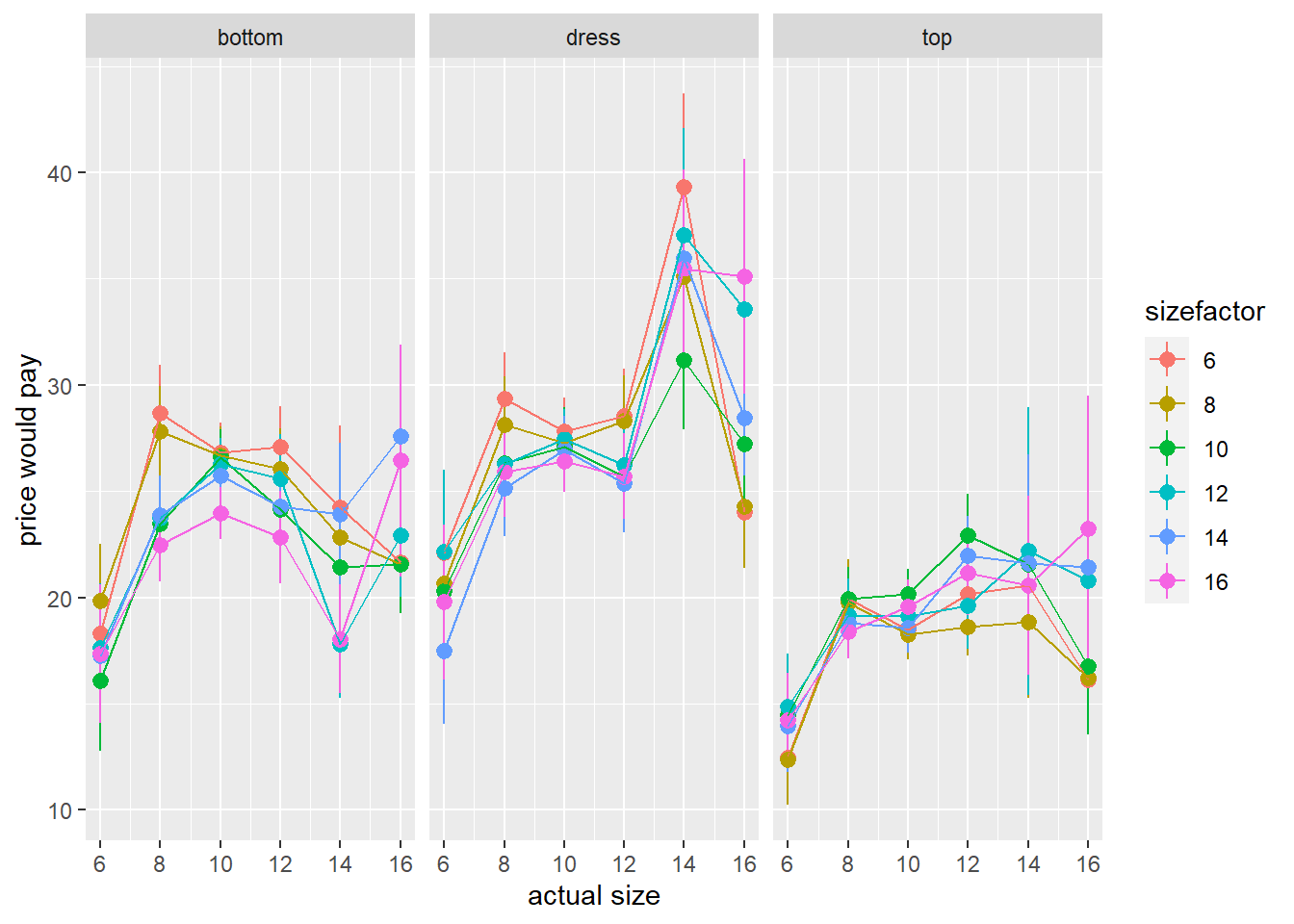
This looks a lot like we are seeing difference, averaged across all participants, in the price they would pay for different types of garment, but not a lot of difference by size.
We can also calculate the ICC’s
Code
library(lme4)
m0 <- lmer(price ~ 1 +
(1|id) + (1|item),
data = df_analysis)
ICC <- as.data.frame(VarCorr(m0))
For participant:
Code
round((ICC[1,4]/sum(ICC[,4]))*100, 2)
And item:
Code
round((ICC[2,4]/sum(ICC[,4]))*100, 2)
So we can see we have a lot of between person variation, and some between item variation in our data. This suggests that there is potential value in our level 2 predictors concerning both the items and between person characteristics.
Visualizations
There are too many ways to visualize this data! Here are just a few:
Code
ggplot(df_analysis, aes(x=c.size, y=price,col=sizefactor))+
stat_summary(geom="pointrange")+
stat_summary(geom="path")+
facet_grid(~item)+
labs(x="actual size",y="price would pay")
Code
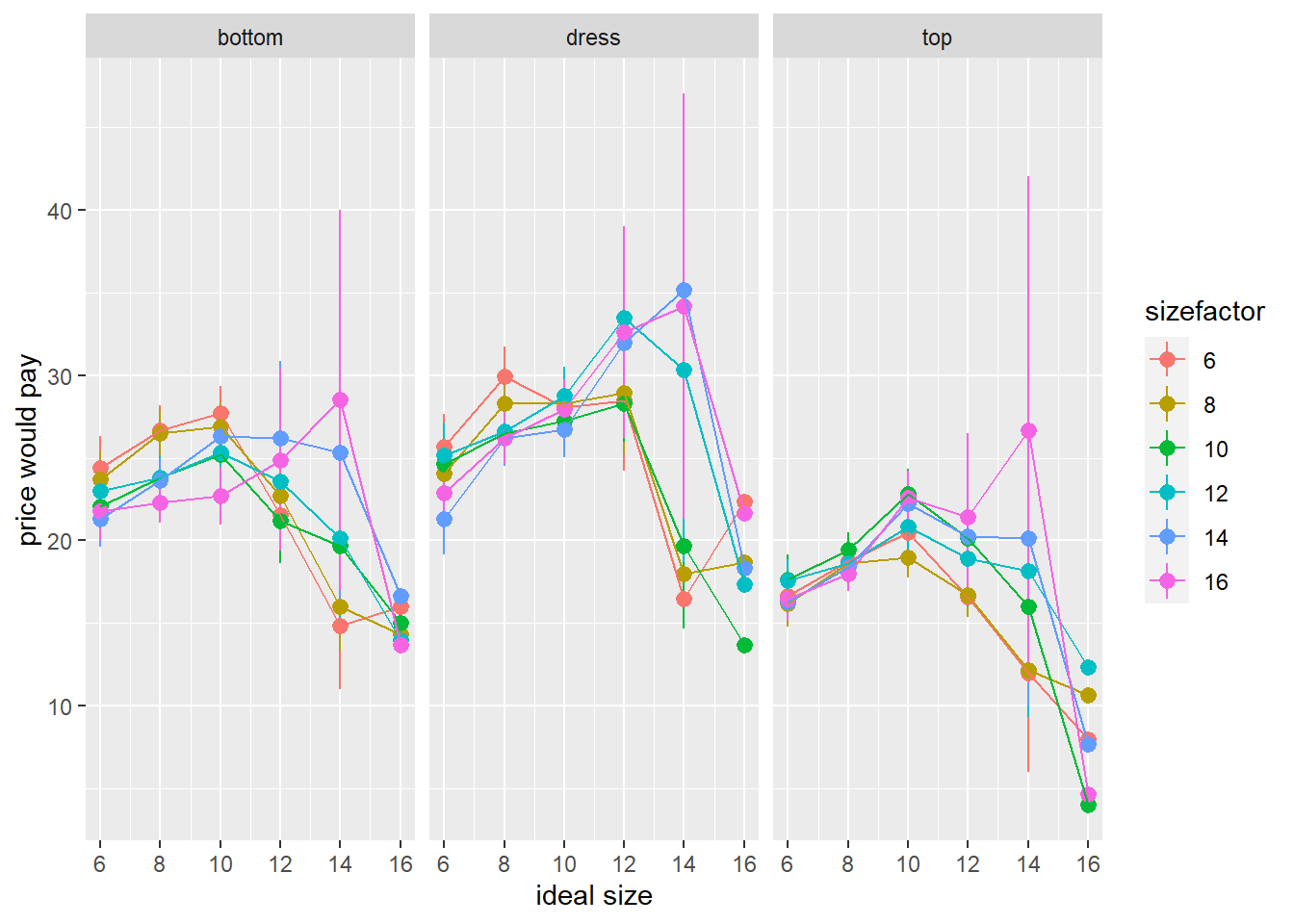
ggplot(df_analysis, aes(x=i.size, y=price,col=sizefactor))+
stat_summary(geom="pointrange")+
stat_summary(geom="path")+
facet_grid(~item)+
labs(x="ideal size",y="price would pay")
Remember, however, that we had some research questions to consider!
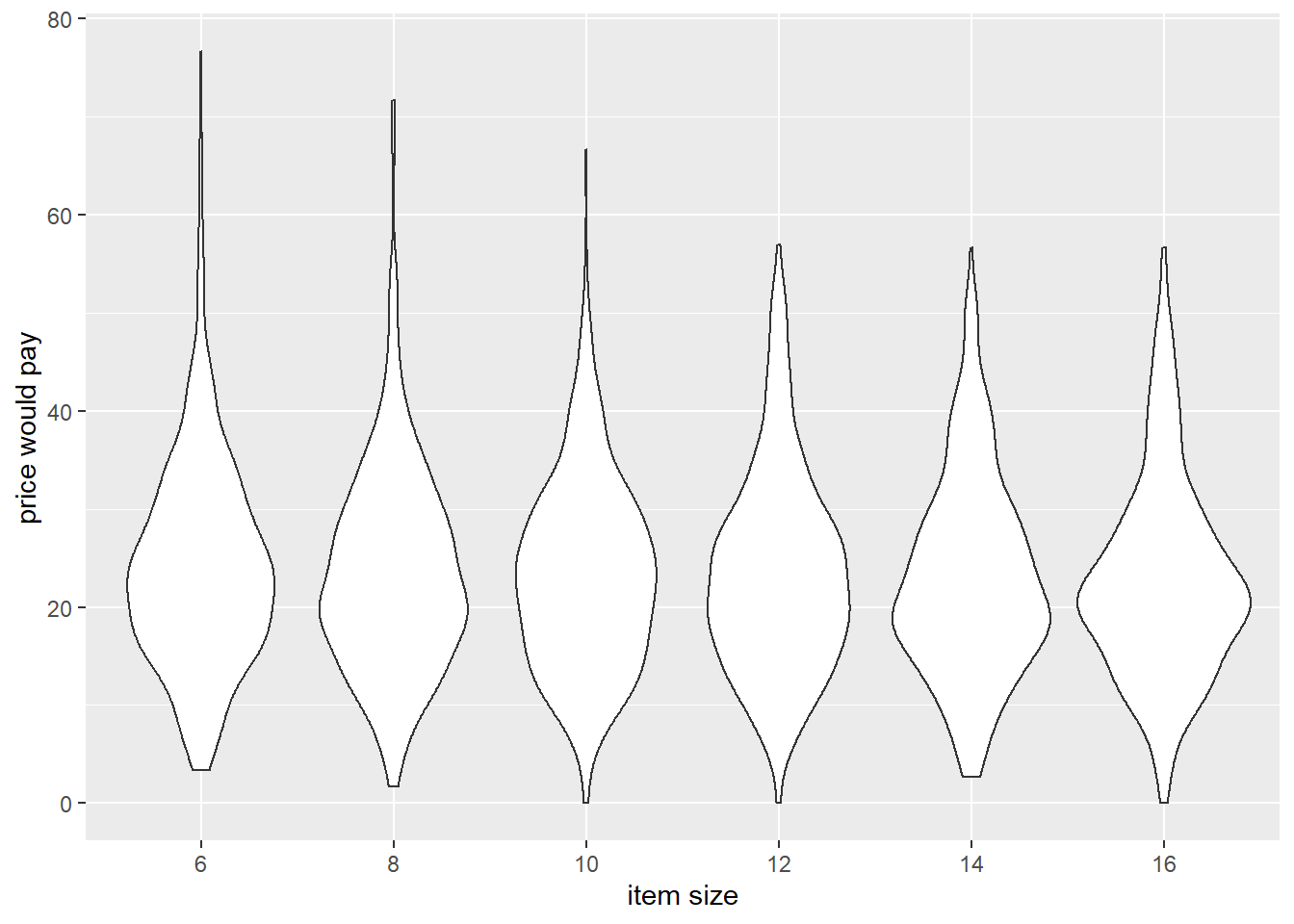
- participants would pay more for garments on thinner models
Code
ggplot(df_analysis, aes(x=sizefactor, y=price))+
geom_violin()+
labs(x="item size",y="price would pay")
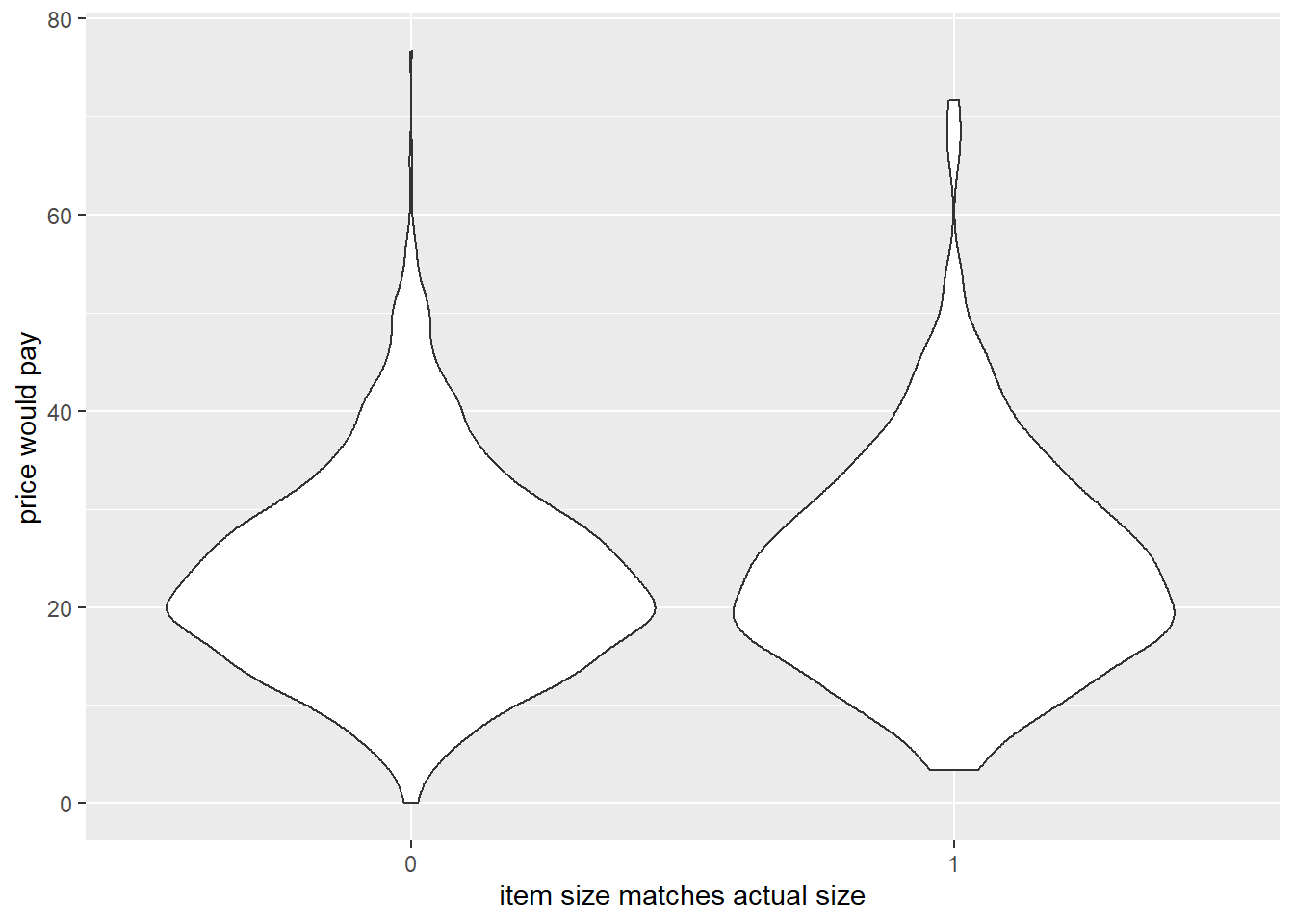
- participants would pay more for items when the model size matched their actual size
Code
ggplot(df_analysis, aes(x=factor(c.match), y=price))+
geom_violin()+
labs(x="item size matches actual size",y="price would pay")
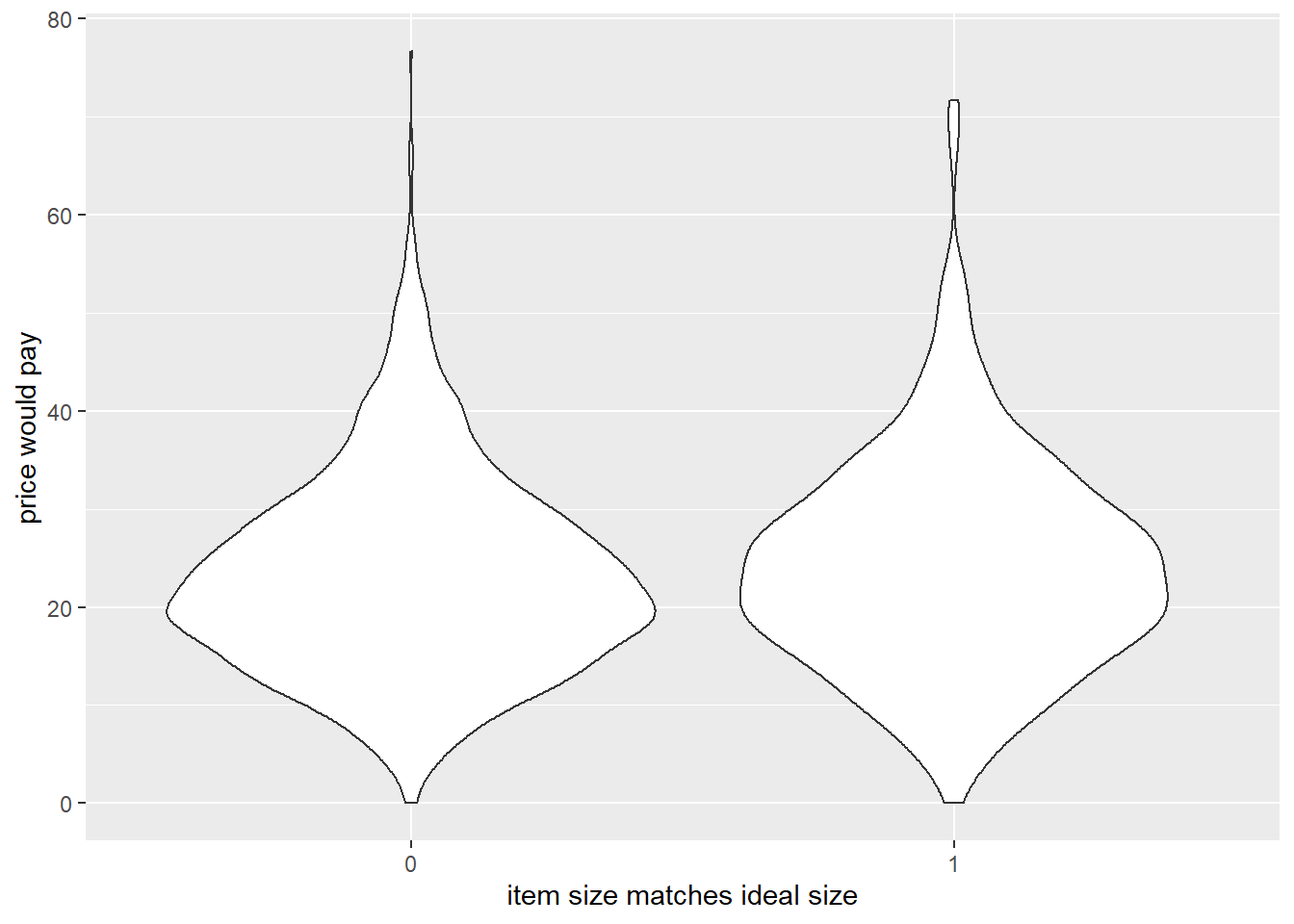
- participants would pay more for items when the model size matched their ideal size
Code
ggplot(df_analysis, aes(x=factor(i.match), y=price))+
geom_violin()+
labs(x="item size matches ideal size",y="price would pay")
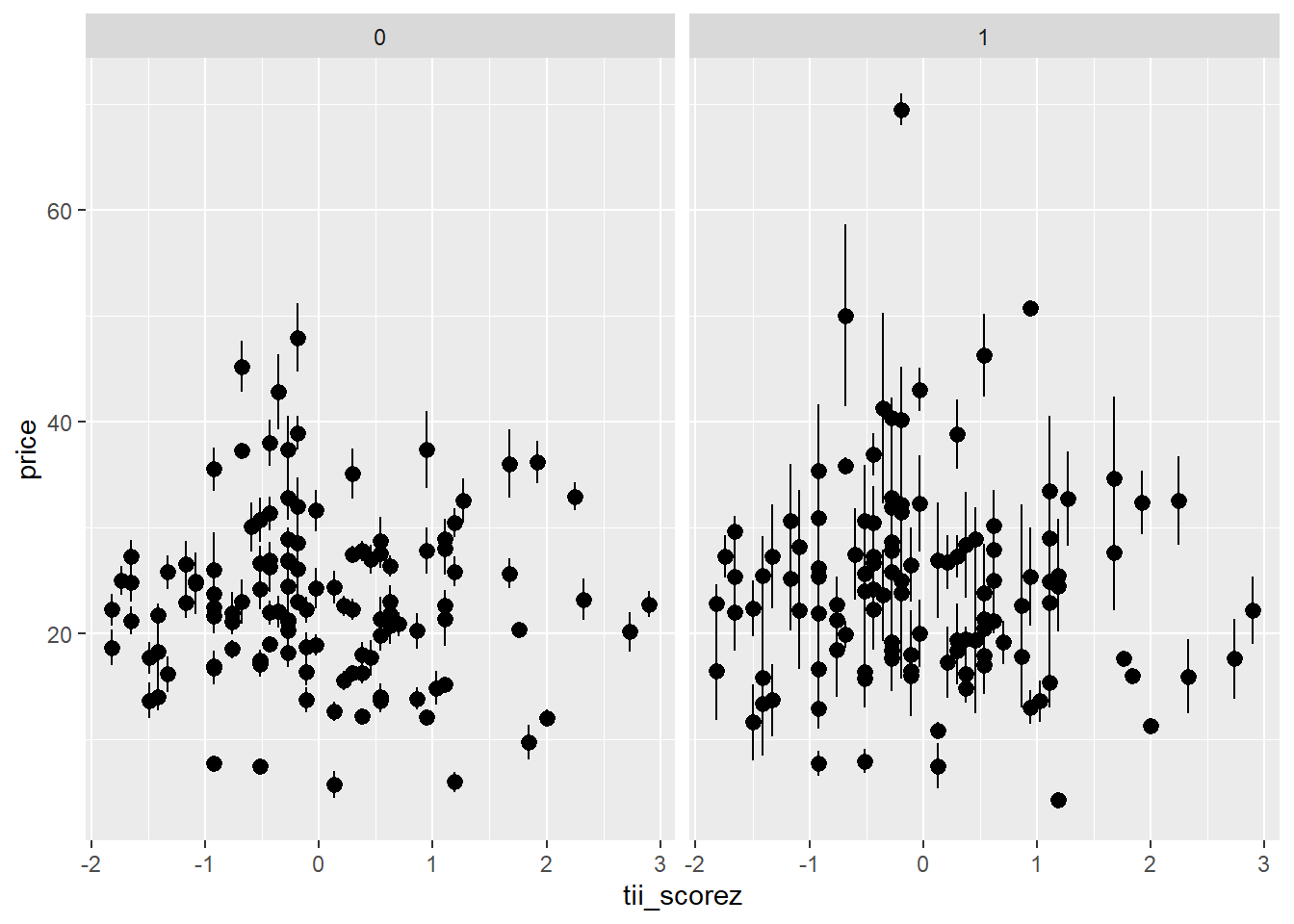
- the extent to which participants idealize thin figures would moderate (ii)
Code
ggplot(df_analysis, aes(x=tii_scorez, y=price))+
stat_summary(geom="pointrange",aes(group=id))+ # want a mean per participant, rather than each individual point
facet_grid(~c.match)
Code
labs(x="thinness idealization score",y="price would pay")
$x
[1] "thinness idealization score"
$y
[1] "price would pay"
attr(,"class")
[1] "labels"
Analysis
Equations
Why item as a fixed effect and not by-item random intercepts (e.g.(1 | item))? It is debatable how you decide to treat item here. It could be argued that the different item types (dress/top/bottom, etc) represent a sample from some broader population of garment types, and as such should be modeled as random effects (especially given that we are not interested in specific parameter estimates for differences between item types).
However, we only have 3 different types of item, and it is important to remember that having (1|item) will involve estimating variance components based on these. Variance estimates will be less stable with such a small number of groups. There’s no hard rule to follow here about “how many groups is enough” (some people suggest 5 or 6 at least), but personally I would be inclined to use item as a fixed effect.
We should also consider the possibility that certain items should be less desirable to purchase when in a smaller/bigger size. It may be that size differences are more salient, or more influential on our outcome variable, for certain types of item over others. This would involve adding the interaction between size and item type
$$
\[\begin{aligned}
\operatorname{price}_{i[j]} =& \beta_{0i} +
\beta_{1i}(\operatorname{item-size}_j) +
\beta_{1}(\operatorname{item-type}_j) + \\
& \beta_{2}(\operatorname{item-size}_j \times \operatorname{item-type}_j) + \\
& \beta_{3i}(\operatorname{matches-actual-size}_j) + \\
& \beta_{4}(\operatorname{matches-ideal-size}_j) + \varepsilon_{i[j]}\\
\qquad \\
\beta_{0i} &= \gamma_{00} + \gamma_{01}(\operatorname{thin-idealisation-score}_i) + \zeta_{0i} \\
\beta_{1i} &= \gamma_{10} + \zeta_{1i}\\
\beta_{3i} &= \gamma_{30} + \gamma_{31}(\operatorname{thin-idealisation-score}_i)\\
& \text{for participant i = 1, } \dots \text{, I} \\
\end{aligned}\]
$$
Fitting the models
We’re going to want to do lots of model comparisons, and for those we need all our models to be fitted on the same data. We’re going to use these variables:
Code
df_analysis <-
df_analysis %>%
filter(
!is.na(size),
!is.na(item),
!is.na(c.match),
!is.na(i.match),
!is.na(tii_scorez),
!is.na(id),
!is.na(price)
)
First, the effect of size. We’re going to treat size as a numeric variable here, in part because it makes our models less complex. It also allows us to model by-participant random effects of size (you can think of this as modeling participants as varying in the amount to which size influences the amount that they are prepared to pay).
Code
m0 <- lmer(price ~ 1 + item +
(1+size|id),
data = df_analysis)
m1 <- lmer(price ~ 1 + item + size +
(1 + size|id),
data = df_analysis)
summary(m1)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item + size + (1 + size | id)
Data: df_analysis
REML criterion at convergence: 14045.7
Scaled residuals:
Min 1Q Median 3Q Max
-4.5003 -0.5885 -0.0396 0.5259 5.1435
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.130 7.882
size 2.371 1.540 -0.07
Residual 33.570 5.794
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.3845 0.7517 32.439
itemdress 2.6468 0.3081 8.590
itemtop -5.3352 0.3073 -17.360
size -0.4893 0.1887 -2.593
Correlation of Fixed Effects:
(Intr) itmdrs itemtp
itemdress -0.204
itemtop -0.205 0.500
size -0.051 0.000 0.001
Code
Data: df_analysis
Models:
m0: price ~ 1 + item + (1 + size | id)
m1: price ~ 1 + item + size + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m0 7 14065 14104 -7025.3 14051
m1 8 14060 14105 -7022.0 14044 6.6082 1 0.01015 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
The interaction between size and item:
Code
m2 <- lmer(price ~ 1 + item * size +
(1+size|id),
data = df_analysis)
summary(m2)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item * size + (1 + size | id)
Data: df_analysis
REML criterion at convergence: 14026.4
Scaled residuals:
Min 1Q Median 3Q Max
-4.5043 -0.5804 -0.0377 0.5095 5.1433
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.135 7.883
size 2.392 1.547 -0.07
Residual 33.240 5.765
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.3870 0.7514 32.454
itemdress 2.6449 0.3066 8.626
itemtop -5.3370 0.3058 -17.451
size -1.1062 0.2587 -4.277
itemdress:size 0.4740 0.3071 1.543
itemtop:size 1.3689 0.3058 4.477
Correlation of Fixed Effects:
(Intr) itmdrs itemtp size itmdr:
itemdress -0.203
itemtop -0.204 0.500
size -0.038 0.002 0.002
itemdrss:sz 0.001 -0.003 -0.002 -0.591
itemtop:siz 0.001 -0.002 -0.002 -0.594 0.500
Code
Data: df_analysis
Models:
m1: price ~ 1 + item + size + (1 + size | id)
m2: price ~ 1 + item * size + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m1 8 14060 14105 -7022.0 14044
m2 10 14043 14100 -7011.7 14023 20.613 2 3.341e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
What about the effect of item size matching your current size:
Code
m3 <- lmer(price ~ 1 + item * size + c.match +
(1+size|id),
data = df_analysis)
summary(m3)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item * size + c.match + (1 + size | id)
Data: df_analysis
REML criterion at convergence: 14023.1
Scaled residuals:
Min 1Q Median 3Q Max
-4.4754 -0.5995 -0.0312 0.5075 5.1647
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.145 7.883
size 2.317 1.522 -0.08
Residual 33.233 5.765
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.2782 0.7537 32.214
itemdress 2.6431 0.3066 8.621
itemtop -5.3374 0.3058 -17.454
size -1.0747 0.2580 -4.166
c.match1 0.6620 0.3494 1.895
itemdress:size 0.4736 0.3071 1.542
itemtop:size 1.3689 0.3058 4.477
Correlation of Fixed Effects:
(Intr) itmdrs itemtp size c.mtc1 itmdr:
itemdress -0.202
itemtop -0.203 0.500
size -0.048 0.002 0.002
c.match1 -0.076 -0.003 -0.001 0.064
itemdrss:sz 0.001 -0.003 -0.002 -0.592 0.000
itemtop:siz 0.001 -0.002 -0.002 -0.595 0.000 0.500
Compare models for current size match:
Code
Data: df_analysis
Models:
m2: price ~ 1 + item * size + (1 + size | id)
m3: price ~ 1 + item * size + c.match + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m2 10 14043 14100 -7011.7 14023
m3 11 14042 14104 -7009.9 14020 3.5856 1 0.05828 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Or your ideal size:
Code
m4 <- lmer(price ~ 1 + item * size + c.match + i.match +
(1+size|id),
data = df_analysis)
summary(m4)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item * size + c.match + i.match + (1 + size | id)
Data: df_analysis
REML criterion at convergence: 14021.9
Scaled residuals:
Min 1Q Median 3Q Max
-4.4705 -0.5911 -0.0355 0.5049 5.1828
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.171 7.885
size 2.281 1.510 -0.08
Residual 33.244 5.766
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.2167 0.7556 32.050
itemdress 2.6439 0.3066 8.622
itemtop -5.3369 0.3058 -17.450
size -1.0264 0.2606 -3.938
c.match1 0.5972 0.3539 1.688
i.match1 0.4308 0.3650 1.180
itemdress:size 0.4736 0.3071 1.542
itemtop:size 1.3685 0.3058 4.475
Correlation of Fixed Effects:
(Intr) itmdrs itemtp size c.mtc1 i.mtc1 itmdr:
itemdress -0.202
itemtop -0.203 0.500
size -0.058 0.002 0.002
c.match1 -0.064 -0.003 -0.001 0.038
i.match1 -0.069 0.002 0.001 0.156 -0.160
itemdrss:sz 0.001 -0.003 -0.002 -0.586 -0.001 0.000
itemtop:siz 0.001 -0.002 -0.002 -0.589 0.000 -0.001 0.500
optimizer (nloptwrap) convergence code: 0 (OK)
Model failed to converge with max|grad| = 0.00383895 (tol = 0.002, component 1)
Compare models for ideal size match:
Code
Data: df_analysis
Models:
m3: price ~ 1 + item * size + c.match + (1 + size | id)
m4: price ~ 1 + item * size + c.match + i.match + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m3 11 14042 14104 -7009.9 14020
m4 12 14042 14110 -7009.2 14018 1.399 1 0.2369
Fixed effects for thin ideals:
Code
m5 <- lmer(price ~ 1 + item * size + c.match + i.match + tii_scorez +
(1+size|id),
data = df_analysis)
summary(m5)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item * size + c.match + i.match + tii_scorez + (1 +
size | id)
Data: df_analysis
REML criterion at convergence: 14020.7
Scaled residuals:
Min 1Q Median 3Q Max
-4.4719 -0.5907 -0.0357 0.5059 5.1829
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.695 7.918
size 2.279 1.510 -0.08
Residual 33.245 5.766
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.21667 0.75848 31.928
itemdress 2.64389 0.30664 8.622
itemtop -5.33684 0.30584 -17.450
size -1.02641 0.26060 -3.939
c.match1 0.59704 0.35388 1.687
i.match1 0.43085 0.36497 1.181
tii_scorez -0.07229 0.73246 -0.099
itemdress:size 0.47367 0.30714 1.542
itemtop:size 1.36846 0.30582 4.475
Correlation of Fixed Effects:
(Intr) itmdrs itemtp size c.mtc1 i.mtc1 t_scrz itmdr:
itemdress -0.201
itemtop -0.202 0.500
size -0.058 0.002 0.002
c.match1 -0.064 -0.003 -0.001 0.038
i.match1 -0.068 0.002 0.001 0.156 -0.160
tii_scorez 0.000 0.000 -0.001 0.001 0.003 -0.001
itemdrss:sz 0.001 -0.003 -0.002 -0.587 -0.001 0.000 0.000
itemtop:siz 0.001 -0.002 -0.002 -0.589 0.000 -0.001 0.000 0.500
Compare models for thin ideal:
Code
Data: df_analysis
Models:
m4: price ~ 1 + item * size + c.match + i.match + (1 + size | id)
m5: price ~ 1 + item * size + c.match + i.match + tii_scorez + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m4 12 14042 14110 -7009.2 14018
m5 13 14044 14118 -7009.2 14018 0.0099 1 0.9208
Do they interact?
Code
m6 <- lmer(price ~ 1 + item * size + c.match + i.match + tii_scorez + c.match*tii_scorez +
(1+size|id),
data = df_analysis)
summary(m6)
Linear mixed model fit by REML ['lmerMod']
Formula: price ~ 1 + item * size + c.match + i.match + tii_scorez + c.match *
tii_scorez + (1 + size | id)
Data: df_analysis
REML criterion at convergence: 14019.8
Scaled residuals:
Min 1Q Median 3Q Max
-4.4672 -0.5966 -0.0323 0.5021 5.1910
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 62.679 7.917
size 2.281 1.510 -0.08
Residual 33.243 5.766
Number of obs: 2130, groups: id, 120
Fixed effects:
Estimate Std. Error t value
(Intercept) 24.20683 0.75845 31.916
itemdress 2.64336 0.30663 8.621
itemtop -5.33608 0.30584 -17.448
size -1.02537 0.26062 -3.934
c.match1 0.56650 0.35506 1.595
i.match1 0.51739 0.37448 1.382
tii_scorez -0.01563 0.73458 -0.021
itemdress:size 0.47423 0.30713 1.544
itemtop:size 1.36867 0.30581 4.476
c.match1:tii_scorez -0.37423 0.36191 -1.034
Correlation of Fixed Effects:
(Intr) itmdrs itemtp size c.mtc1 i.mtc1 t_scrz itmdr: itmtp:
itemdress -0.201
itemtop -0.202 0.500
size -0.056 0.002 0.002
c.match1 -0.063 -0.003 -0.001 0.037
i.match1 -0.070 0.002 0.002 0.153 -0.173
tii_scorez -0.001 0.000 0.000 0.001 -0.003 0.016
itemdrss:sz 0.001 -0.003 -0.002 -0.586 -0.001 0.001 0.000
itemtop:siz 0.001 -0.002 -0.002 -0.589 0.000 -0.001 0.000 0.500
c.mtch1:t_s 0.013 0.002 -0.002 -0.004 0.082 -0.224 -0.076 -0.002 -0.001
And let’s just compare all at once, just for fun:
Code
anova(m0,m1,m2,m3,m4,m5,m6)
Data: df_analysis
Models:
m0: price ~ 1 + item + (1 + size | id)
m1: price ~ 1 + item + size + (1 + size | id)
m2: price ~ 1 + item * size + (1 + size | id)
m3: price ~ 1 + item * size + c.match + (1 + size | id)
m4: price ~ 1 + item * size + c.match + i.match + (1 + size | id)
m5: price ~ 1 + item * size + c.match + i.match + tii_scorez + (1 + size | id)
m6: price ~ 1 + item * size + c.match + i.match + tii_scorez + c.match * tii_scorez + (1 + size | id)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
m0 7 14065 14104 -7025.3 14051
m1 8 14060 14105 -7022.0 14044 6.6082 1 0.01015 *
m2 10 14043 14100 -7011.7 14023 20.6131 2 3.341e-05 ***
m3 11 14042 14104 -7009.9 14020 3.5856 1 0.05828 .
m4 12 14042 14110 -7009.2 14018 1.3990 1 0.23690
m5 13 14044 14118 -7009.2 14018 0.0099 1 0.92076
m6 14 14045 14125 -7008.7 14017 1.0720 1 0.30048
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Check model(s)
Code
library(lattice)
library(gridExtra)
grid.arrange(
plot(m1, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
plot(m2, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
plot(m3, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
plot(m4, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
plot(m5, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
plot(m6, sqrt(abs(resid(.)))~fitted(.), type = c("p","smooth")),
ncol=3
)
Code
grid.arrange(
qqmath(m1),
qqmath(m2),
qqmath(m3),
qqmath(m4),
qqmath(m5),
qqmath(m6),
ncol=3
)
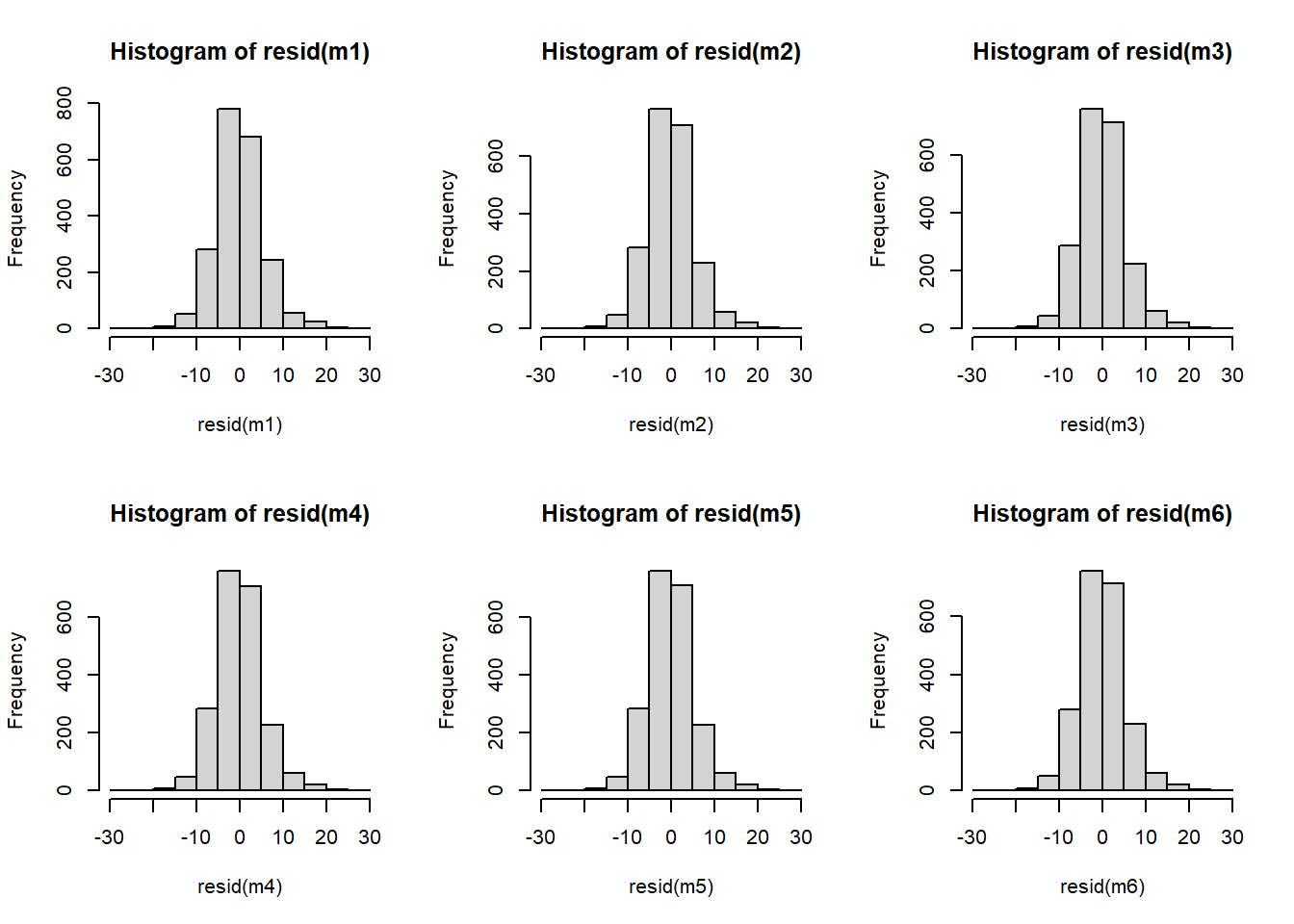
I might be a little concerned about our assumptions here. The QQplots look a little off in the tails, however, the histograms of residuals make it look okay:
For peace of mind, we could re-do the model comparisons with parametric bootstrapped model comparison for more reliable conclusions. This will assume that the model specified distribution of residuals (\(\varepsilon \sim N(0, \sigma_\varepsilon)\)) holds in the population
Code
library(pbkrtest)
PBmodcomp(m1, m0)
stat df p.value
LRT 6.605558 1 0.01016610
PBtest 6.605558 NA 0.01098901
stat df p.value
LRT 20.61468 2 3.338708e-05
PBtest 20.61468 NA 9.990010e-04
stat df p.value
LRT 3.585967 1 0.05826951
PBtest 3.585967 NA 0.07092907
stat df p.value
LRT 1.399085 1 0.2368768
PBtest 1.399085 NA 0.2317682
stat df p.value
LRT 0.00153228 1 0.9687753
PBtest 0.00153228 NA 0.9670330
stat df p.value
LRT 1.072739 1 0.3003275
PBtest 1.072739 NA 0.2857143
Visualise model(s)
Code
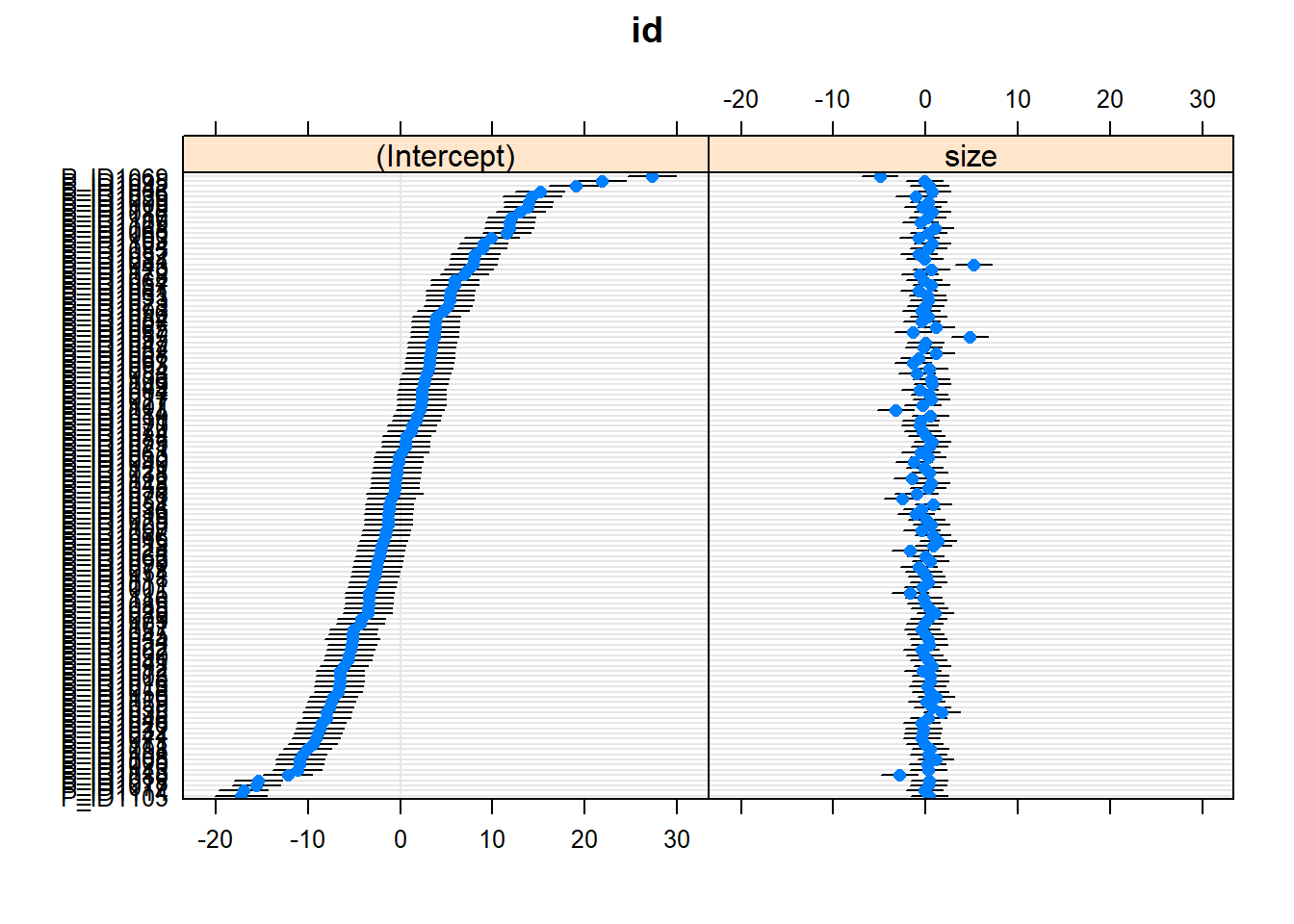
dotplot.ranef.mer(ranef(m6))
Code
library(sjPlot)
plot_model(m6, type = "pred")$size
Code
plot_model(m6, type = "pred")$c.match
Code
plot_model(m6, type = "pred")$i.match
Code
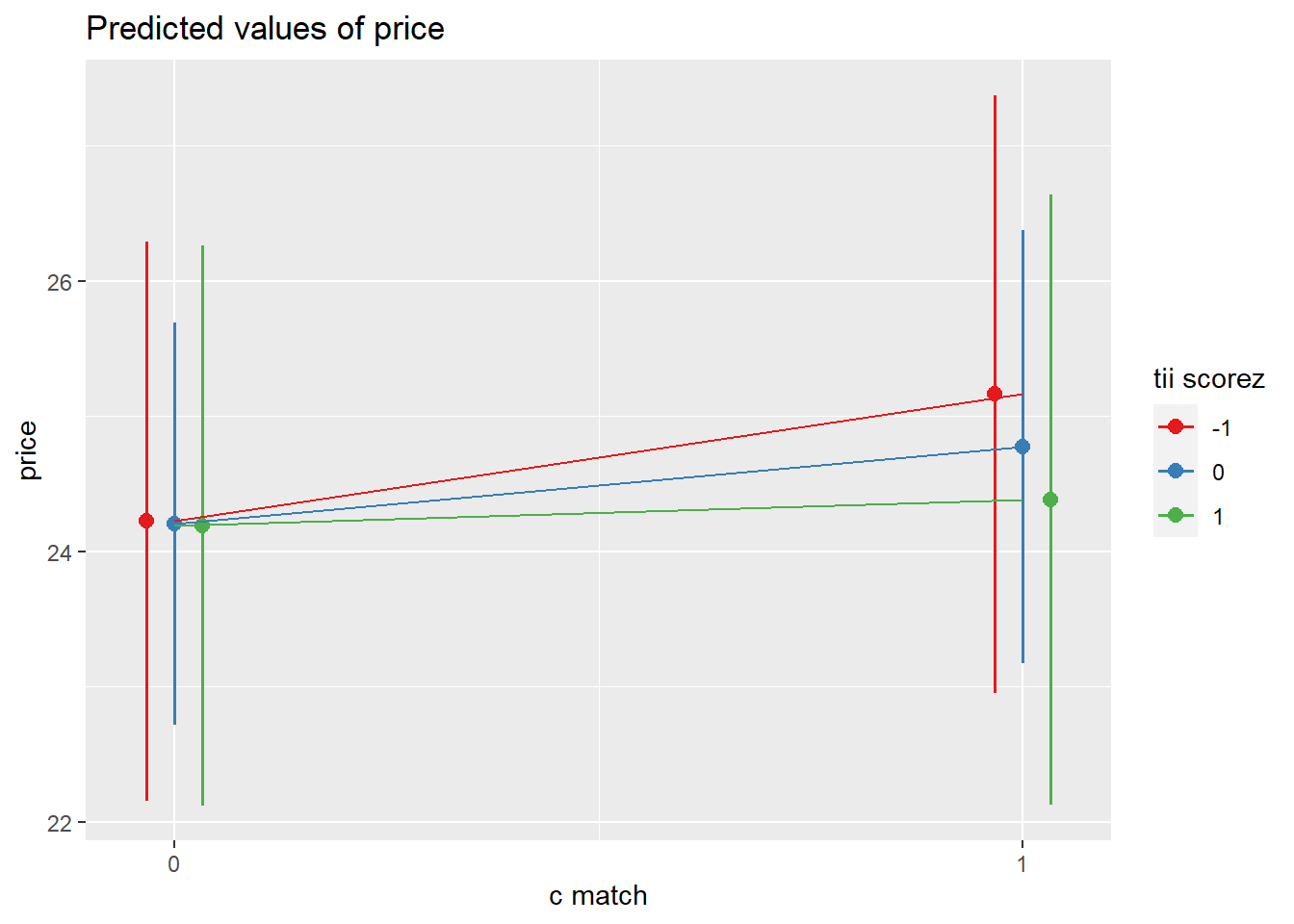
plot_model(m6, type = "pred", terms = c("c.match","tii_scorez [-1,0,1]")) +
geom_path()
Interpret model(s)
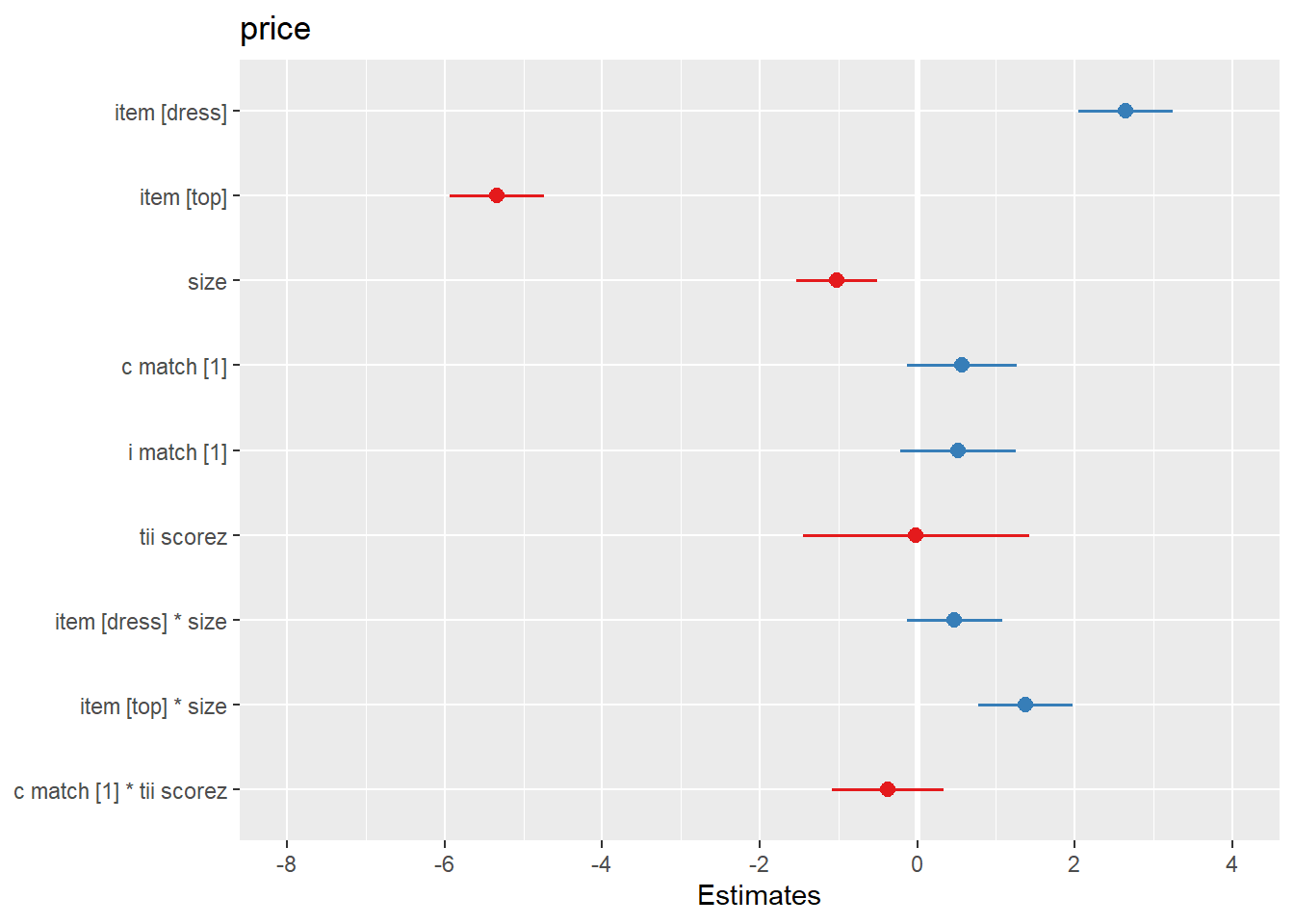
The amount that participants indicated they would pay for a garment was modeled using multi-level linear regression models, with by-participant random intercepts and effects of garment size. After adjusting for effects on our outcome due to the garment-type (dress/top/bottom), inclusion of garment size as a fixed predictor was found to improve model fit (Parametric Bootstrap Likelihood Ratio = 6.61, p = 0.011). The effect of garment-size was found to differ between garment-types (inclusion of interaction term LRT 20.61, p <.001). After controlling for garment-type, -size and their interaction, neither the garments’ matching participants’ actual or ideal size, nor the extent to which participants idealized thin figures were found to predict the price they would pay, and the predicted interaction between actual-size matching and idealization of thin figures was not evidenced. Results of the full model are shown in Table 1.
Code
tab_model(m6, show.p = F, title="Table 1: Model results. Please note that 95% CIs are not bootstrapped")
Table 1: Model results. Please note that 95% CIs are not bootstrapped
| |
price |
| Predictors |
Estimates |
CI |
| (Intercept) |
24.21 |
22.72 – 25.69 |
| item [dress] |
2.64 |
2.04 – 3.24 |
| item [top] |
-5.34 |
-5.94 – -4.74 |
| size |
-1.03 |
-1.54 – -0.51 |
| c match [1] |
0.57 |
-0.13 – 1.26 |
| i match [1] |
0.52 |
-0.22 – 1.25 |
| tii scorez |
-0.02 |
-1.46 – 1.42 |
| item [dress] * size |
0.47 |
-0.13 – 1.08 |
| item [top] * size |
1.37 |
0.77 – 1.97 |
| c match [1] * tii scorez |
-0.37 |
-1.08 – 0.34 |
| Random Effects |
| σ2 |
33.24 |
| τ00 id |
62.68 |
| τ11 id.size |
2.28 |
| ρ01 id |
-0.08 |
| ICC |
0.66 |
| N id |
120 |
| Observations |
2130 |
| Marginal R2 / Conditional R2 |
0.106 / 0.697 |