Each of these pages provides an analysis run through for a different type of design. Each document is structured in the same way:
- First the data and research context is introduced. For the purpose of these tutorials, we will only use examples where the data can be shared - either because it is from an open access publication, or because it is unpublished or simulated.
- Second, we go through any tidying of the data that is required, before creating some brief descriptives and visualizations of the raw data.
- Then, we conduct an analysis. Where possible, we translate the research questions into formal equations prior to fitting the models in lme4. Model comparisons are conducted, along with checks of distributional assumptions on our model residuals.
- Finally, we visualize and interpret our analysis.
Please note that there will be only minimal explanation of the steps undertaken here, as these pages are intended as example analyses rather than additional labs readings. Please also be aware that there are many decisions to be made throughout conducting analyses, and it may be the case that you disagree with some of the choices we make here. As always with these things, it is how we justify our choices that is important. We warmly welcome any feedback and suggestions to improve these examples: please email ug.ppls.stats@ed.ac.uk.
Overview
Intervention studies are where the researchers intervenes (e.g. through administration of a drug or treatment) as part of the study
The data used for this worked example are simulated to represent data from 50 participants, each measured at 3 different time-points (pre, during, and post) on a measure of stress. Half of the participants are in the control group, and half are in the treatment group (e.g. half received some cognitive behavioural therapy (CBT), half did not).
Code
set.seed(983)
library(tidyverse)
simMIX <- tibble(
ppt = factor(rep(paste("ID", 1:50, sep=""),each=3)),
ppt_int = rep(rnorm(50,0,15), each=3),
stress = round(
c(rnorm(75,c(50,45,42),sd=c(5, 6, 5)),
rnorm(75,c(55,30,20),sd=c(6, 10, 5))) + ppt_int,
0),
time = as_factor(rep(c("Pre", "During", "Post"), 50)),
group = as_factor(c(rep("Control", 75), rep("Treatment", 75)))
) %>% select(-ppt_int)
Data Wrangling
Because we simulated our data, it is already nice and tidy. Each observation is a row, and we have variable indicating participant identifier (the ppt variable).
Code
ppt stress time group
ID1 : 3 Min. :-13.00 Pre :50 Control :75
ID10 : 3 1st Qu.: 29.00 During:50 Treatment:75
ID11 : 3 Median : 43.00 Post :50
ID12 : 3 Mean : 42.22
ID13 : 3 3rd Qu.: 55.75
ID14 : 3 Max. : 90.00
(Other):132
Code
bind_rows(head(simMIX), tail(simMIX))
# A tibble: 12 × 4
ppt stress time group
<fct> <dbl> <fct> <fct>
1 ID1 65 Pre Control
2 ID1 81 During Control
3 ID1 55 Post Control
4 ID2 62 Pre Control
5 ID2 57 During Control
6 ID2 67 Post Control
7 ID49 58 Pre Treatment
8 ID49 34 During Treatment
9 ID49 25 Post Treatment
10 ID50 35 Pre Treatment
11 ID50 21 During Treatment
12 ID50 0 Post Treatment
Descriptives
We now have two factors of interest, scores over Time and by Group. So we do our descriptive statistics by multiple grouping factors.
Code
sumMIX <-
simMIX %>%
group_by(time, group) %>%
summarise(
N = n_distinct(ppt),
Mean = round(mean(stress, na.rm=T),2),
SD = round(sd(stress, na.rm=T),2)
)
Code
library(kableExtra)
kable(sumMIX) %>%
kable_styling("striped")
| time |
group |
N |
Mean |
SD |
| Pre |
Control |
25 |
52.84 |
14.60 |
| Pre |
Treatment |
25 |
54.76 |
16.78 |
| During |
Control |
25 |
49.64 |
13.23 |
| During |
Treatment |
25 |
30.44 |
15.49 |
| Post |
Control |
25 |
46.08 |
14.24 |
| Post |
Treatment |
25 |
19.56 |
15.99 |
Visualizations
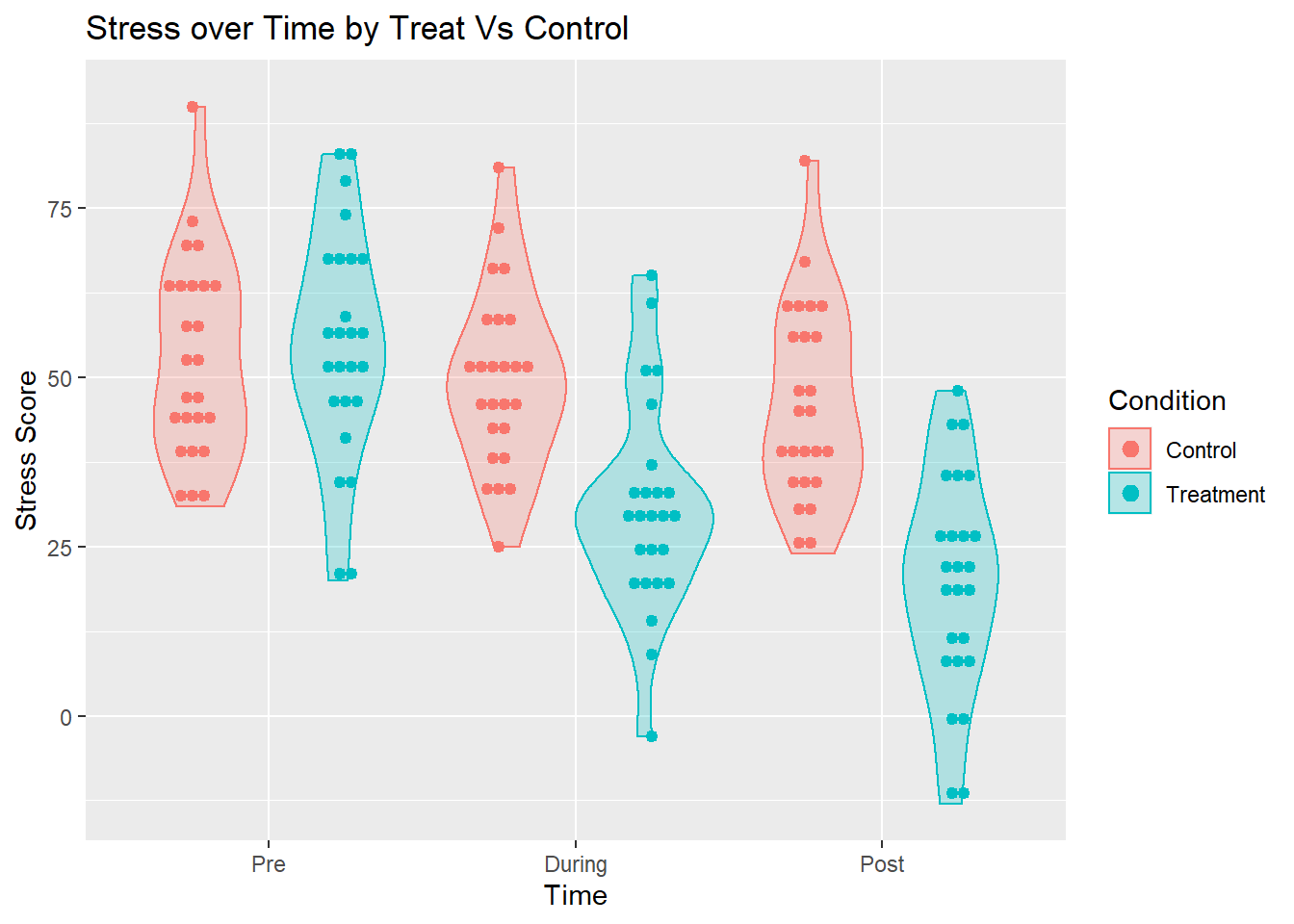
We can use a violin plot to visualize our data.
Code
simMIX %>%
ggplot(aes(x=time, y=stress, color=group, fill=group)) +
geom_violin(alpha = .25) +
geom_dotplot(binaxis='y', stackdir='center', position=position_dodge(1),
dotsize = 0.5) +
labs(x="Time", y = "Stress Score",
title="Stress over Time by Treat Vs Control", color="Condition", fill="Condition")
It is useful to note that the violin plot contains much of the same information that a more traditional boxplot would have, but provides more information on the actual distribution with groups as it draws on density plots.
What we can see here is that over time, the control group shows little change, but the treatment group shows distinct declines.
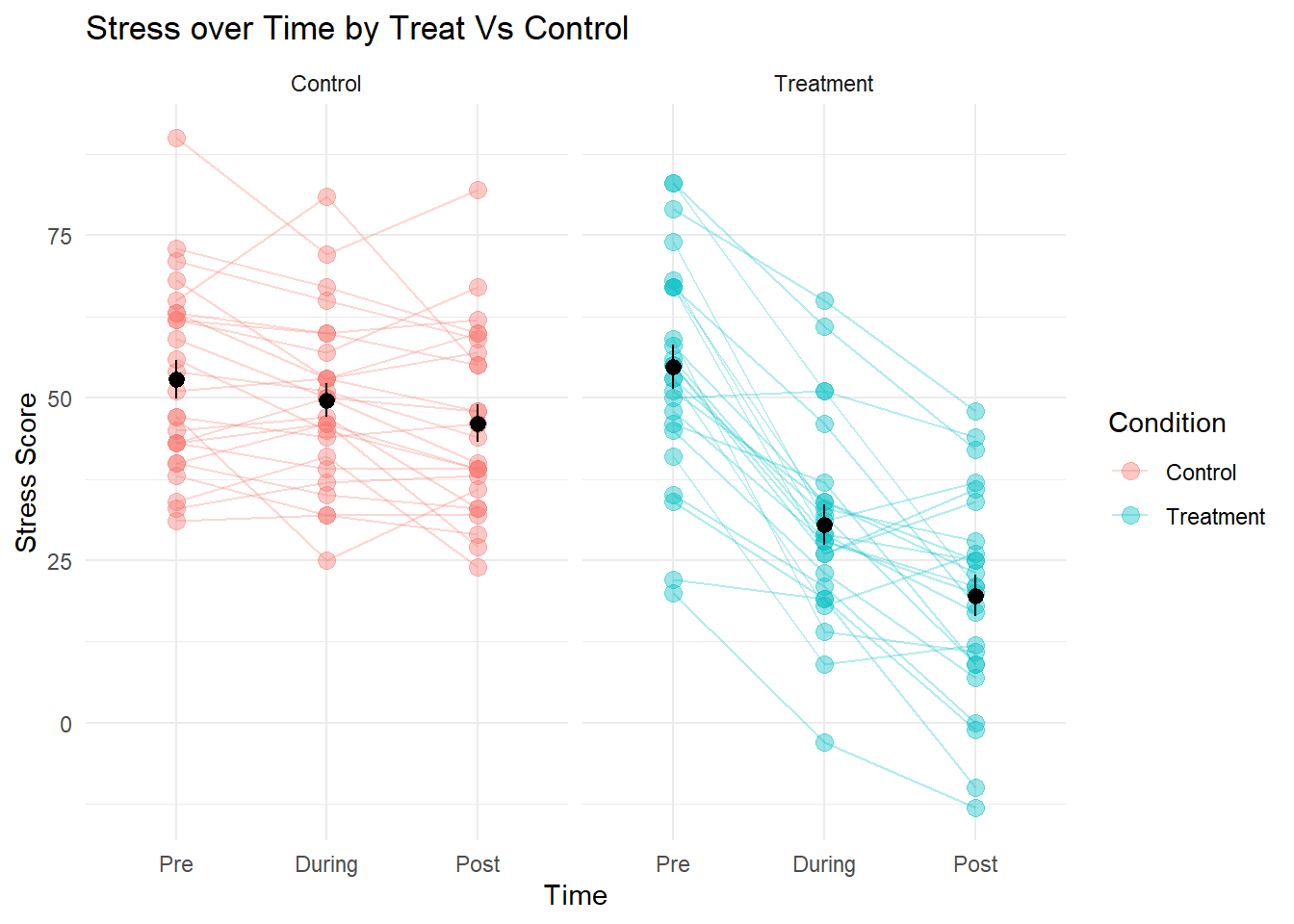
We can also consider line plots with separate facets for each condition:
Code
simMIX %>%
ggplot(aes(x=time, y=stress, col=group)) +
geom_point(size=3, alpha=.4)+
geom_line(aes(group=ppt), alpha=.3)+
stat_summary(geom="pointrange", col="black")+
facet_wrap(~group)+
labs(x="Time", y = "Stress Score",
title="Stress over Time by Treat Vs Control", color="Condition", fill="Condition")+
theme_minimal()
Analysis
Equations
\[
\begin{aligned}
\operatorname{stress}_{i[j]} &= \beta_{0i} + \beta_{1i}(\operatorname{timeDuring}_j) + \beta_{2i}(\operatorname{timePost}_j) + \varepsilon_{i[j]} \\
\beta_{0i} &= \gamma_{00} + \gamma_{01}(\operatorname{groupTreatment}_i) + \zeta_{0i} \\
\beta_{1i} &= \gamma_{10} + \gamma_{11}(\operatorname{groupTreatment}_i) \\
\beta_{2i} &= \gamma_{20} + \gamma_{21}(\operatorname{groupTreatment}_i) \\
& \text{for ppt i = 1,} \dots \text{, I}
\end{aligned}
\]
Fitting the models
Base model:
Code
m0 <- lmer(stress ~ 1 +
(1 | ppt), data = simMIX)
summary(m0)
Linear mixed model fit by REML ['lmerMod']
Formula: stress ~ 1 + (1 | ppt)
Data: simMIX
REML criterion at convergence: 1286.7
Scaled residuals:
Min 1Q Median 3Q Max
-2.02099 -0.49670 0.01511 0.49445 1.96548
Random effects:
Groups Name Variance Std.Dev.
ppt (Intercept) 179.7 13.40
Residual 209.7 14.48
Number of obs: 150, groups: ppt, 50
Fixed effects:
Estimate Std. Error t value
(Intercept) 42.220 2.234 18.9
Main effects:
Code
m1 <- lmer(stress ~ 1 + time + group +
(1 | ppt), data = simMIX)
summary(m1)
Linear mixed model fit by REML ['lmerMod']
Formula: stress ~ 1 + time + group + (1 | ppt)
Data: simMIX
REML criterion at convergence: 1185.9
Scaled residuals:
Min 1Q Median 3Q Max
-1.69666 -0.62649 0.01574 0.59245 1.92387
Random effects:
Groups Name Variance Std.Dev.
ppt (Intercept) 166.57 12.91
Residual 98.01 9.90
Number of obs: 150, groups: ppt, 50
Fixed effects:
Estimate Std. Error t value
(Intercept) 61.100 3.046 20.061
timeDuring -13.760 1.980 -6.950
timePost -20.980 1.980 -10.596
groupTreatment -14.600 3.992 -3.657
Correlation of Fixed Effects:
(Intr) tmDrng timPst
timeDuring -0.325
timePost -0.325 0.500
groupTrtmnt -0.655 0.000 0.000
Interaction:
Code
m2 <- lmer(stress ~ 1 + time + group + time*group +
(1 | ppt), data = simMIX)
summary(m2)
Linear mixed model fit by REML ['lmerMod']
Formula: stress ~ 1 + time + group + time * group + (1 | ppt)
Data: simMIX
REML criterion at convergence: 1096.5
Scaled residuals:
Min 1Q Median 3Q Max
-1.83946 -0.53326 -0.05639 0.53766 2.31546
Random effects:
Groups Name Variance Std.Dev.
ppt (Intercept) 184.82 13.595
Residual 43.26 6.577
Number of obs: 150, groups: ppt, 50
Fixed effects:
Estimate Std. Error t value
(Intercept) 52.840 3.020 17.494
timeDuring -3.200 1.860 -1.720
timePost -6.760 1.860 -3.634
groupTreatment 1.920 4.272 0.449
timeDuring:groupTreatment -21.120 2.631 -8.028
timePost:groupTreatment -28.440 2.631 -10.810
Correlation of Fixed Effects:
(Intr) tmDrng timPst grpTrt tmDr:T
timeDuring -0.308
timePost -0.308 0.500
groupTrtmnt -0.707 0.218 0.218
tmDrng:grpT 0.218 -0.707 -0.354 -0.308
tmPst:grpTr 0.218 -0.354 -0.707 -0.308 0.500
Comparison with aov()
As we did with the simple repeated measures, we can also compare the sums of squares breakdown for the LMM (m2) by calling anova() on the lmer() model.
First with aov():
Code
m3 <- aov(stress ~ time + group + time*group +
Error(ppt), data = simMIX)
summary(m3)
Error: ppt
Df Sum Sq Mean Sq F value Pr(>F)
group 1 7993 7993 13.37 0.000633 ***
Residuals 48 28691 598
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
time 2 11360 5680 131.31 <2e-16 ***
time:group 2 5452 2726 63.01 <2e-16 ***
Residuals 96 4153 43
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
And then summarise:
Code
Analysis of Variance Table
npar Sum Sq Mean Sq F value
time 2 11360.4 5680.2 131.305
group 1 578.5 578.5 13.373
time:group 2 5452.0 2726.0 63.014
For ease, lets compare all models with a parametric bootstrap likelihood ratio test:
Code
library(pbkrtest)
PBmodcomp(m1, m0)
PBmodcomp(m2, m1)
Bootstrap test; time: 58.26 sec; samples: 1000; extremes: 0;
large : stress ~ 1 + time + group + (1 | ppt)
stress ~ 1 + (1 | ppt)
stat df p.value
LRT 90.348 3 < 2.2e-16 ***
PBtest 90.348 0.000999 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Bootstrap test; time: 60.30 sec; samples: 1000; extremes: 0;
large : stress ~ 1 + time + group + time * group + (1 | ppt)
stress ~ 1 + time + group + (1 | ppt)
stat df p.value
LRT 83.853 2 < 2.2e-16 ***
PBtest 83.853 0.000999 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
And extract some bootstrap 95% CIs
Code
confint(m2,method="boot")
2.5 % 97.5 %
.sig01 11.063915 16.8989137
.sigma 5.570576 7.3318954
(Intercept) 46.958215 58.9035049
timeDuring -6.598935 -0.1161317
timePost -10.473009 -3.1302747
groupTreatment -7.001190 10.0517531
timeDuring:groupTreatment -26.395306 -16.1101659
timePost:groupTreatment -33.665532 -23.2588741
Check Model
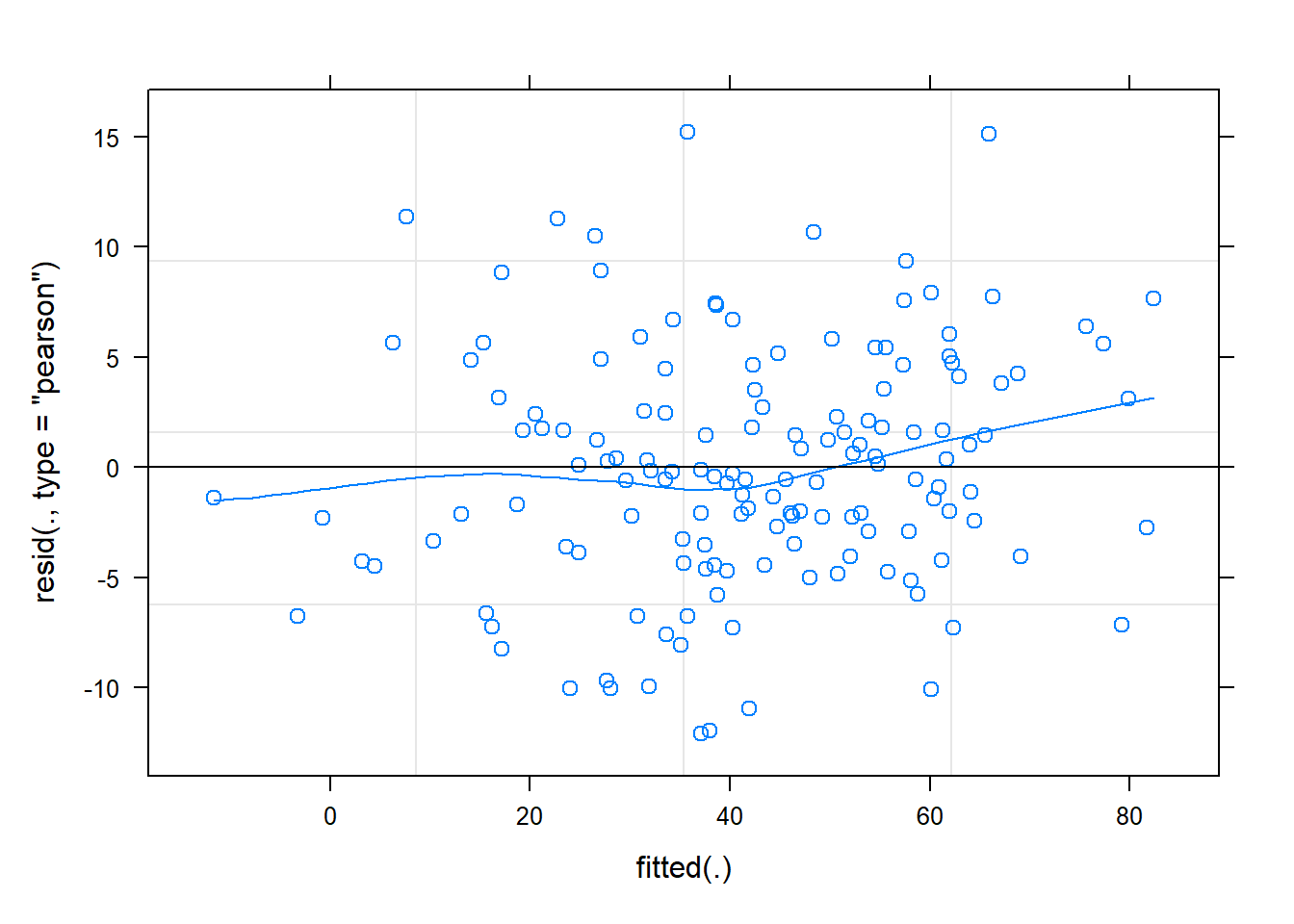
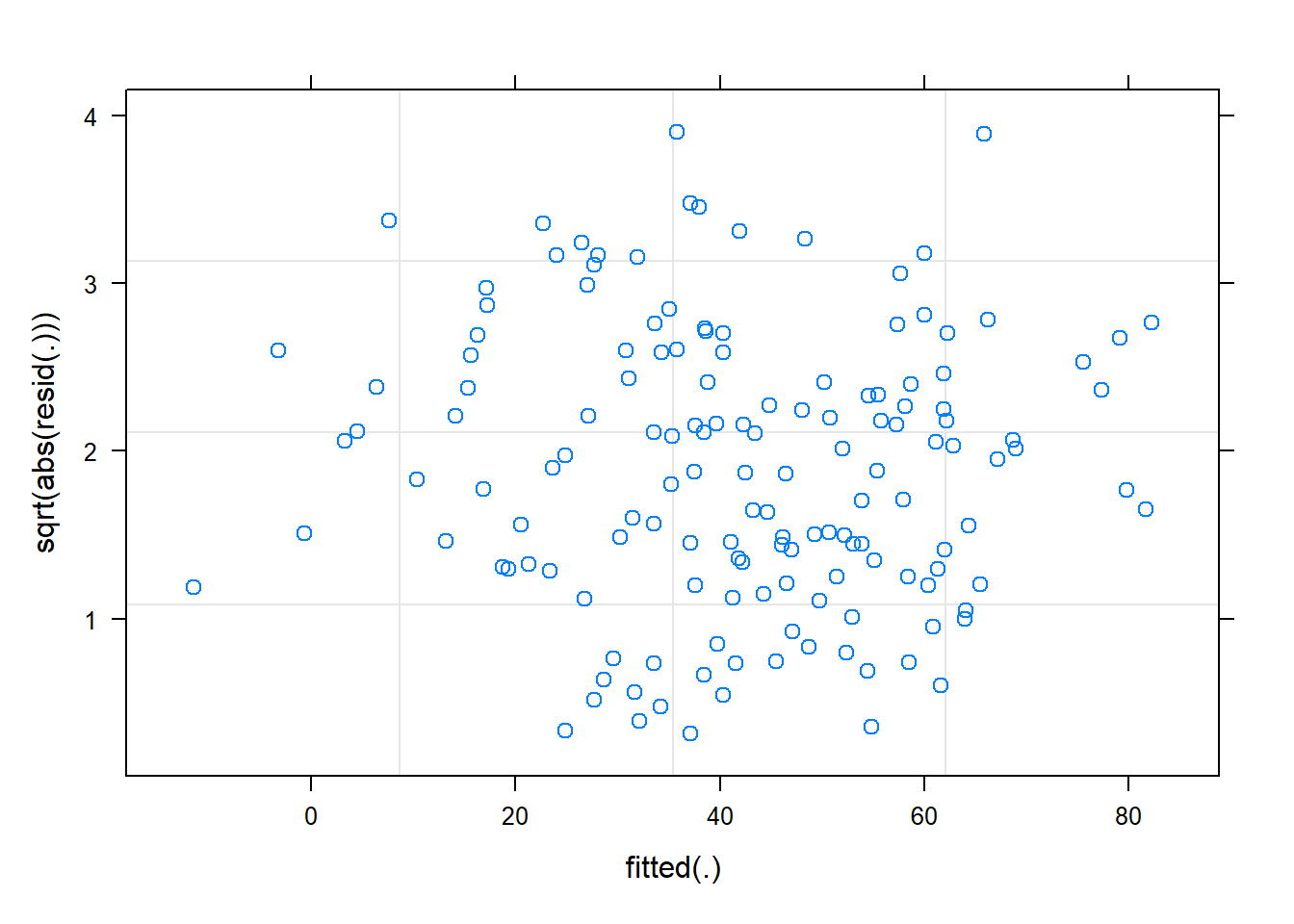
The residuals look reasonably normally distributed, and there seems to be fairly constant variance across the linear predictor. We might be a little concerned about the potential tails of the plot below, at which residuals don’t appear to have a mean of zero
Code
plot(m2, type = c("p","smooth"))
Code
plot(m2, sqrt(abs(resid(.)))~fitted(.))
Code
library(lattice)
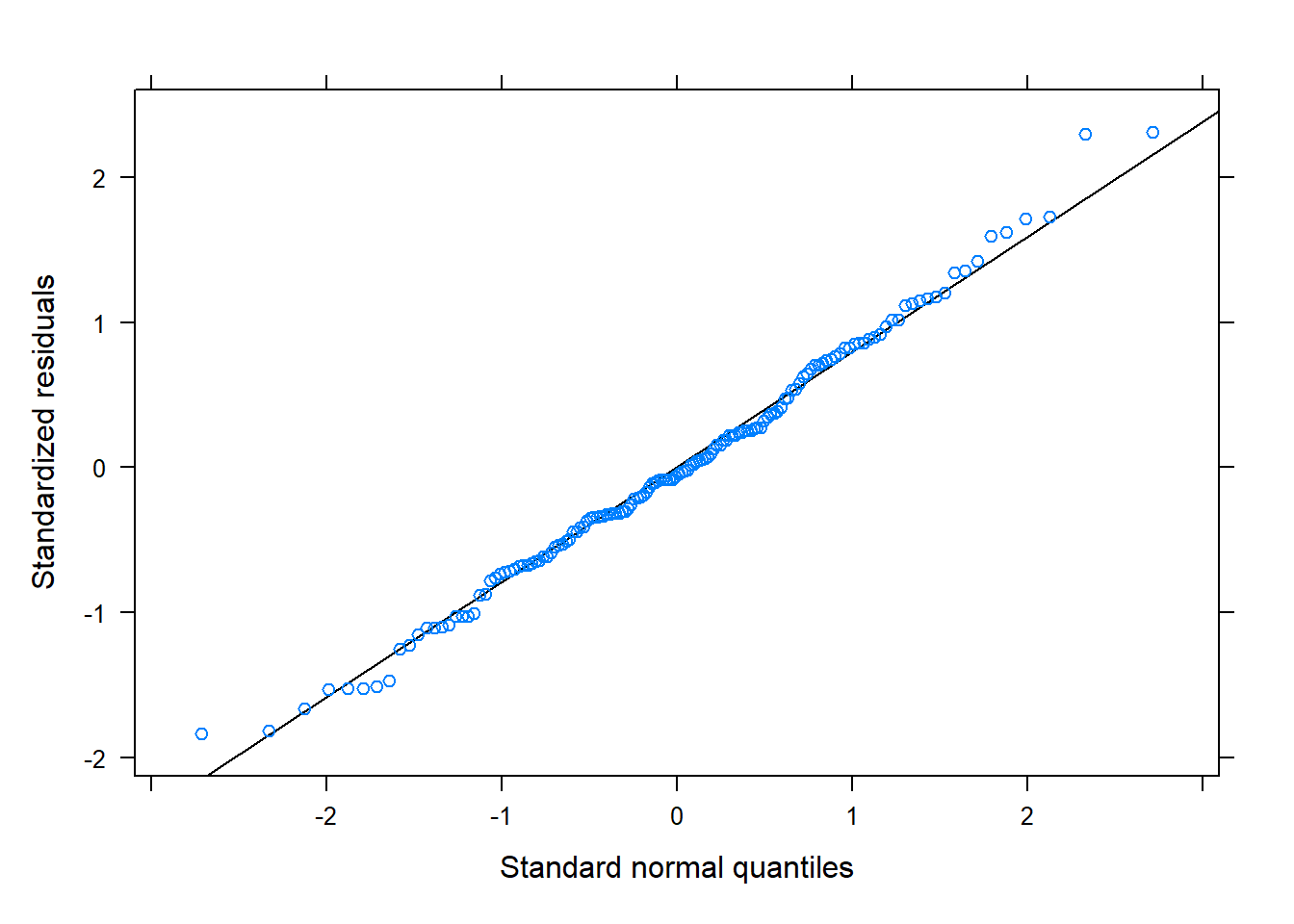
qqmath(m2)
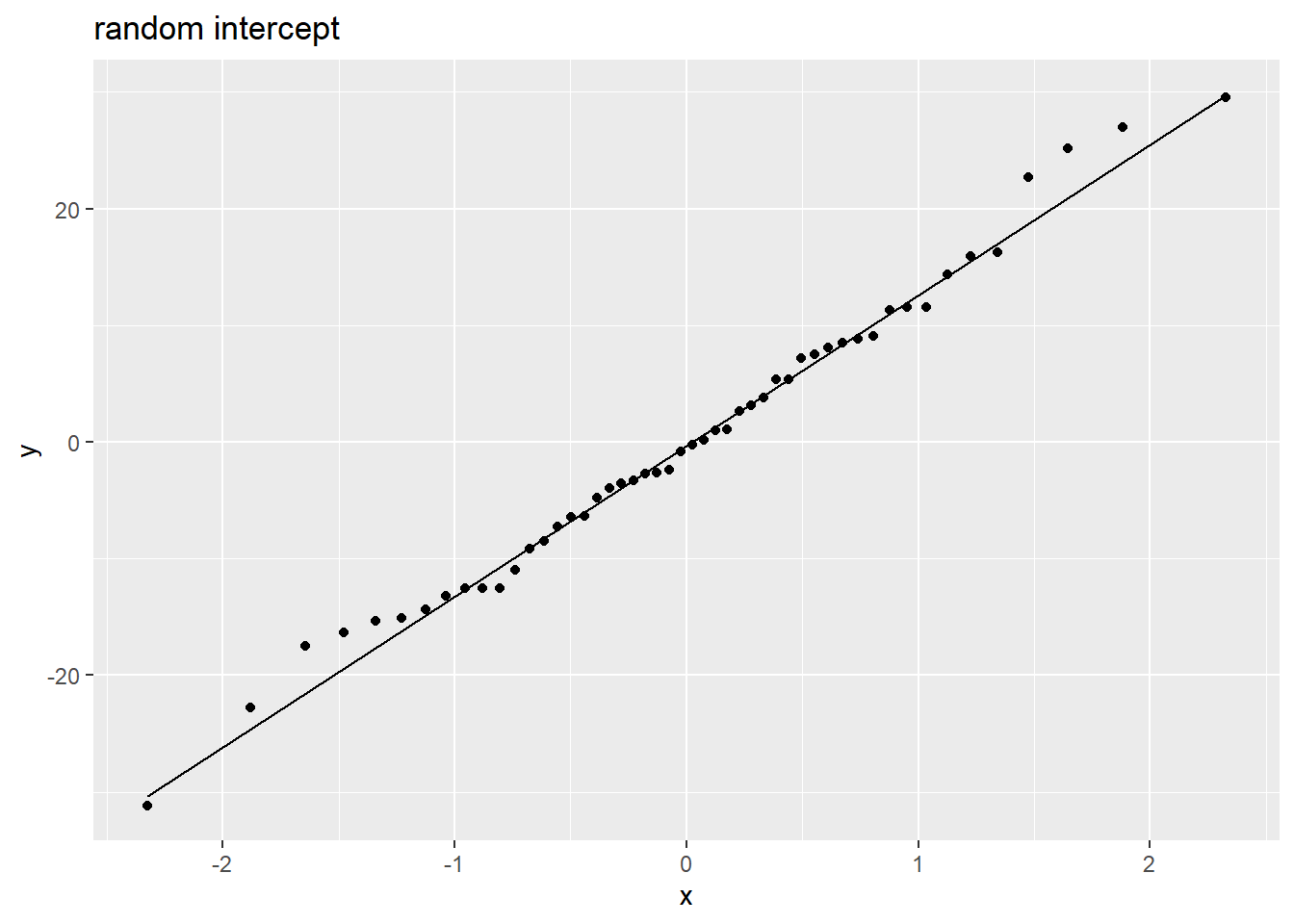
Random effects are (roughly) normally distributed:
Code
rans <- as.data.frame(ranef(m2)$ppt)
ggplot(rans, aes(sample = `(Intercept)`)) +
stat_qq() + stat_qq_line() +
labs(title="random intercept")
Visualise Model
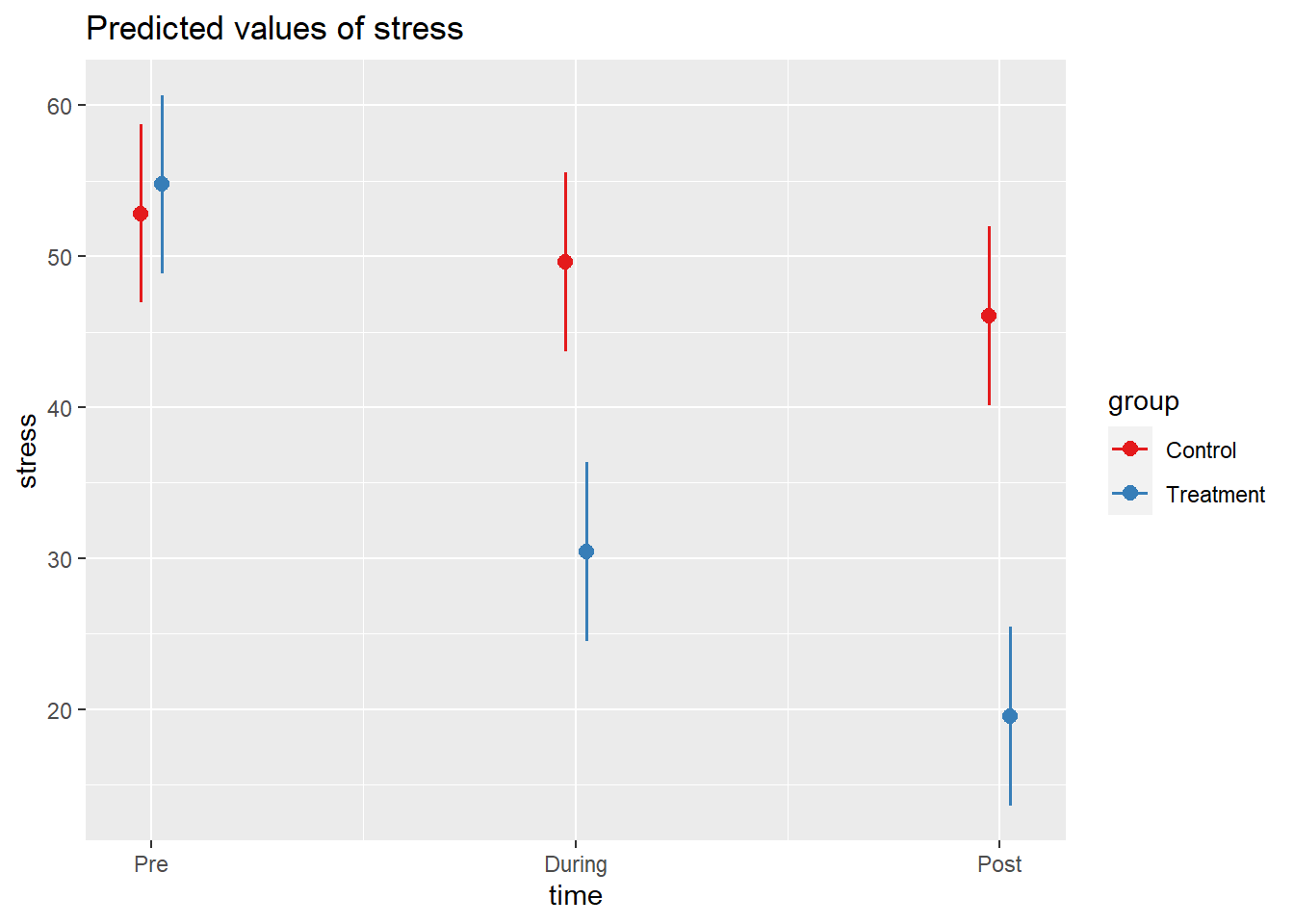
Code
library(sjPlot)
plot_model(m2, type="int")
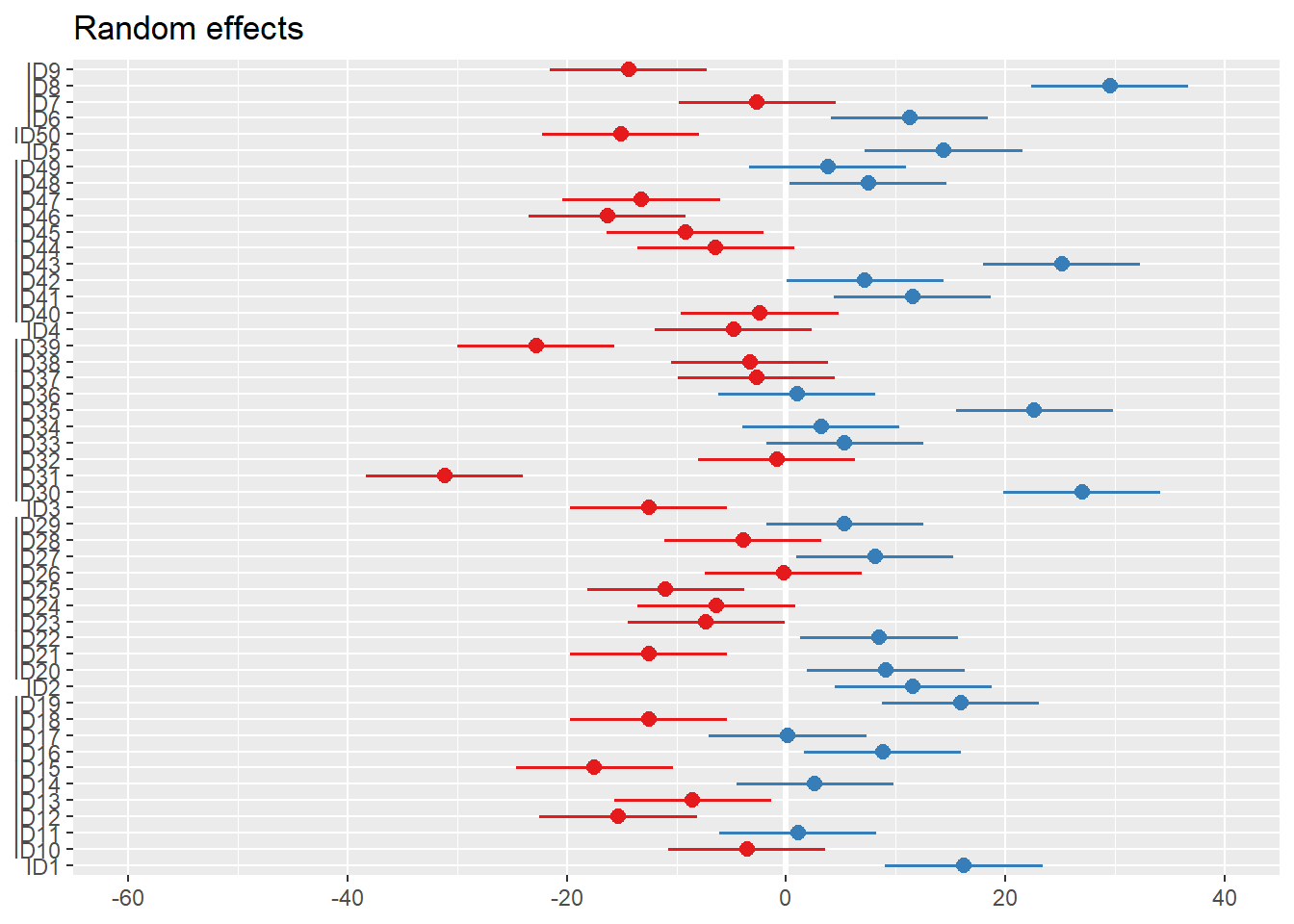
Code
plot_model(m2, type="re") # an alternative: dotplot.ranef.mer(ranef(m1))
Interpret model
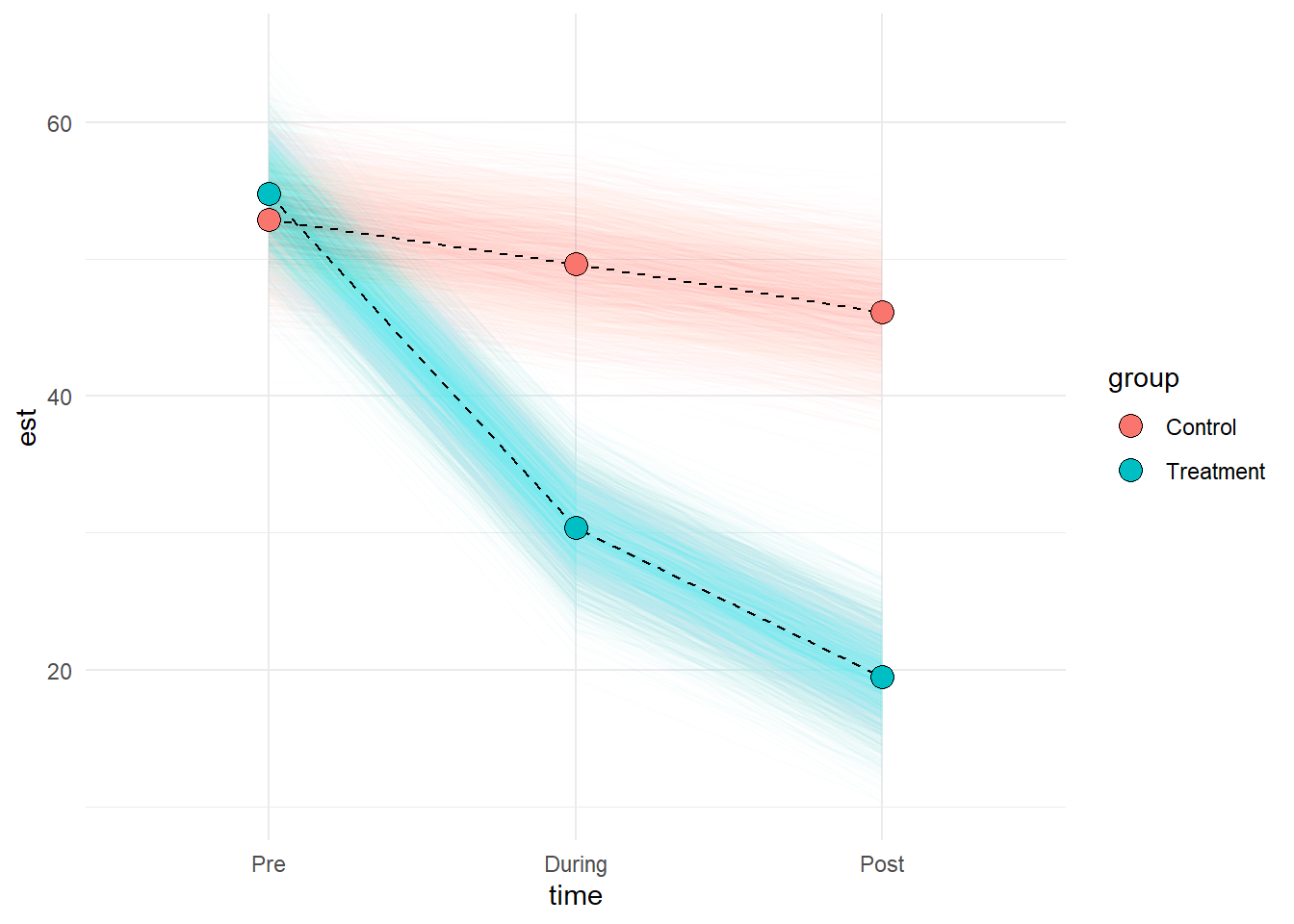
Stress at each study time-point (pre-, during- and post-intervention) was modeled using linear mixed effects models, with by-participant random intercepts, and the incremental addition of time, condition (control vs treatment) and their interaction. An interaction between time and condition improved model fit over the model without the interaction (Parametric bootstrap LRT 83.85, p < .001), suggesting that patterns of change over time in stress levels differed for the treatment group from the control group.
Result show that for the control group, stress did not show clear change between time-points 1 and 2 (-3.2, 95% CI [-6.92 – 0.32]), but by time-point 3 had reduced relative to time-point 1 by -6.76 [-10.58 – -3.3] points. Prior to the intervention (time-point 1), the treatment group did not appear to differ from the control group with respect to their level of stress, but showed an additional -21.12 [-26.75 – -15.63] point change by time-point 2 and -28.44 [-33.75 – -23.23] points by the end of the study (time-point 3). This pattern of results is shown in Figure 1.