Multiple Comparisons
Introduction
This week’s exercises are much lighter than last week, so it might be worth also using it to catch up on previous week’s this term.
Next week :
This week, we’re going to take a brief look at the idea of multiple comparisons.
Refreshing last week:
Last week we learned how to conduct a 2-factor ANOVA. We examined the effects of two variables, Diagnosis (Amnesic, Huntingtons or a Control group) and Task (Grammar, Classification or Recognition).
We incrementally built the 2x2 anova model.
Recall, ANOVA is actually a special case of the linear model in which the predictors are categorical variables, so we can build our model using lm(), and presented slightly differently.
Our full model, including the interaction, looked like this:
mdl_int <- lm(Score ~ 1 + Diagnosis + Task + Diagnosis:Task, data = cog)
# Alternatively, we could have used the following shorter version:
# mdl_int <- lm(Score ~ 1 + Diagnosis * Task, data = cog)And to view the typically presented ANOVA table for this model:
anova(mdl_int)| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| Diagnosis | 2 | 5250 | 2625 | 16.64 | 7.64e-06 |
| Task | 2 | 5250 | 2625 | 16.64 | 7.64e-06 |
| Diagnosis:Task | 4 | 5000 | 1250 | 7.923 | 0.0001092 |
| Residuals | 36 | 5680 | 157.8 | NA | NA |
The interaction between diagnosis and task is significant. At the 5% level, the probability of obtaining an F-statistic as large as 7.92 or larger, if there was no interaction effect, is <.001. This provides very strong evidence against the null hypothesis that effect of task is constant across the different diagnoses.
(remember: in the presence of a significant interaction it does not make sense to interpret the main effects as their interpretation changes with the level of the other factor)
ANOVA is an “omnibus” test
The results from an ANOVA are often called an ‘omnibus’ test, because we are testing the null hypothesis that a set of group means are equal (or in the interaction case, that the differences between group means are equal across some other factor).
If you have found Semester 1 materials on linear models a bit more intuitive than ANOVA, another way to think of it is that we are testing the improvement of model fit between a full and a reduced model.
For instance, our significant interaction \(F(4, 36) = 7.9225, p < .001\) can also be obtained by comparing the nested models (one with the interaction vs one without):
mdl_add <- lm(Score ~ 1 + Diagnosis + Task, data = cog)
mdl_int <- lm(Score ~ 1 + Diagnosis + Task + Diagnosis:Task, data = cog)
anova(mdl_add, mdl_int)## Analysis of Variance Table
##
## Model 1: Score ~ 1 + Diagnosis + Task
## Model 2: Score ~ 1 + Diagnosis + Task + Diagnosis:Task
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 40 10680
## 2 36 5680 4 5000 7.9225 0.0001092 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1But it is common to want to know more about the details of such an effect. What groups differ, and by how much?
The traditional approach is to conduct an ANOVA, and then only ask this sort of follow-up question if obtaining a significant omnibus test.
You might think of it as:
Question 1: Omnibus: “Are there any differences in group means?”
Question 2: Comparisons: “What are the differences and between which groups?”
If your answer to 1 is “No,” then it doesn’t make much sense to ask question 2.
Multiple Comparisons
In last week’s exercises we began to look at how we compare different groups, by using contrast analysis to conduct tests of specific comparisons between groups. We also saw how we might conduct “pairwise comparisons,” where we test all possible pairs of group means within a given set.
For instance, we compares the means of the different diagnosis groups for each task:
emm_task <- emmeans(mdl_int, ~ Diagnosis | Task)
contr_task <- contrast(emm_task, method = 'pairwise',
adjust = "bonferroni")
contr_taskor we might test all different combinations of task and diagnosis group (if that was something we were theoretically interested in, which is unlikely!) which would equate to conducting 36 comparisons!
emm_task <- emmeans(mdl_int, ~ Diagnosis * Task)
contr_task <- contrast(emm_task, method = 'pairwise',
adjust = "bonferroni")
contr_task## contrast estimate SE df t.ratio
## control grammar - amnesic grammar 20 7.94 36 2.518
## control grammar - huntingtons grammar 40 7.94 36 5.035
## control grammar - control classification 0 7.94 36 0.000
## control grammar - amnesic classification 10 7.94 36 1.259
## control grammar - huntingtons classification 35 7.94 36 4.406
## control grammar - control recognition -15 7.94 36 -1.888
## control grammar - amnesic recognition 15 7.94 36 1.888
## control grammar - huntingtons recognition -15 7.94 36 -1.888
## amnesic grammar - huntingtons grammar 20 7.94 36 2.518
## amnesic grammar - control classification -20 7.94 36 -2.518
## amnesic grammar - amnesic classification -10 7.94 36 -1.259
## amnesic grammar - huntingtons classification 15 7.94 36 1.888
## amnesic grammar - control recognition -35 7.94 36 -4.406
## amnesic grammar - amnesic recognition -5 7.94 36 -0.629
## amnesic grammar - huntingtons recognition -35 7.94 36 -4.406
## huntingtons grammar - control classification -40 7.94 36 -5.035
## huntingtons grammar - amnesic classification -30 7.94 36 -3.776
## huntingtons grammar - huntingtons classification -5 7.94 36 -0.629
## huntingtons grammar - control recognition -55 7.94 36 -6.923
## huntingtons grammar - amnesic recognition -25 7.94 36 -3.147
## huntingtons grammar - huntingtons recognition -55 7.94 36 -6.923
## control classification - amnesic classification 10 7.94 36 1.259
## control classification - huntingtons classification 35 7.94 36 4.406
## control classification - control recognition -15 7.94 36 -1.888
## control classification - amnesic recognition 15 7.94 36 1.888
## control classification - huntingtons recognition -15 7.94 36 -1.888
## amnesic classification - huntingtons classification 25 7.94 36 3.147
## amnesic classification - control recognition -25 7.94 36 -3.147
## amnesic classification - amnesic recognition 5 7.94 36 0.629
## amnesic classification - huntingtons recognition -25 7.94 36 -3.147
## huntingtons classification - control recognition -50 7.94 36 -6.294
## huntingtons classification - amnesic recognition -20 7.94 36 -2.518
## huntingtons classification - huntingtons recognition -50 7.94 36 -6.294
## control recognition - amnesic recognition 30 7.94 36 3.776
## control recognition - huntingtons recognition 0 7.94 36 0.000
## amnesic recognition - huntingtons recognition -30 7.94 36 -3.776
## p.value
## 0.5907
## 0.0005
## 1.0000
## 1.0000
## 0.0033
## 1.0000
## 1.0000
## 1.0000
## 0.5907
## 0.5907
## 1.0000
## 1.0000
## 0.0033
## 1.0000
## 0.0033
## 0.0005
## 0.0207
## 1.0000
## <.0001
## 0.1190
## <.0001
## 1.0000
## 0.0033
## 1.0000
## 1.0000
## 1.0000
## 0.1190
## 0.1190
## 1.0000
## 0.1190
## <.0001
## 0.5907
## <.0001
## 0.0207
## 1.0000
## 0.0207
##
## P value adjustment: bonferroni method for 36 tests
Why does the number of tests matter?
As discussed briefly in last week’s exercises, we will ideally ensure that our error rate is 0.05 (i.e., the chance that we reject a null hypothesis when it actually true, is 5%).
But this error-rate applies to each statistical hypothesis we test. So if we conduct an experiment in which we plan on conducting lots of tests of different comparisons, the chance of an error being made increases substantially. Across the family of tests performed that chance will be much higher than 5%.1
Each test conducted at \(\alpha = 0.05\) has a 0.05 (or 5%) probability of Type I error (wrongly rejecting the null hypothesis). If we do 9 tests, that experimentwise error rate is \(\alpha_{ew} \leq 9 \times 0.05\), where 9 is the number of comparisons made as part of the experiment. Thus, if nine independent comparisons were made at the \(\alpha = 0.05\) level, the experimentwise Type I error rate \(\alpha_{ew}\) would be at most \(9 \times 0.05 = 0.45\). That is, we could wrongly reject the null hypothesis on average 45 times out of 100. To make this more confusing, many of the tests in a family are not independent (see the lecture slides for the calculation of error rate for dependent tests).
Here, we go through some of the different options available to us to control, or ‘correct’ for this problem. The first we look at was used in the solutions to last week’s exercises, and is perhaps the most well-known.
Bonferroni
Bonferroni
- Use Bonferroni’s method when you are interested in a small number of planned contrasts (or pairwise comparisons).
- Bonferroni’s method is to divide alpha by the number of tests/confidence intervals.
- Assumes that all comparisons are independent of one another.
- It sacrifices slightly more power than Tukey’s method (discussed below), but it can be applied to any set of contrasts or linear combinations (i.e., it is useful in more situations than Tukey).
- It is usually better than Tukey if we want to do a small number of planned comparisons.
Load the data from last week, and re-acquaint yourself with it.
Provide a plot of the Diagnosis*Task group mean scores.
The data is at https://uoepsy.github.io/data/cognitive_experiment.csv.
Fit the interaction model, using lm().
Pass your model to the anova() function, to remind yourself that there is a significant interaction present.
There are various ways to make nice tables in RMarkdown.
Some of the most well known are:
- The knitr package has
kable()
- The pander package has
pander()
Pick one (or find go googling and find a package you like the look of), install the package (if you don’t already have it), then try to create a nice pretty ANOVA table rather than the one given by anova(model).
As in the previous week’s exercises, let us suppose that we are specifically interested in comparisons of the mean score across the different diagnosis groups for a given task.
Edit the code below to obtain the pairwise comparisons of diagnosis groups for each task. Use the Bonferroni method to adjust for multiple comparisons, and then obtain confidence intervals.
library(emmeans)
emm_task <- emmeans(mdl_int, ? )
contr_task <- contrast(emm_task, method = ?, adjust = ? )
adjusting \(\alpha\), adjusting p
In the lecture we talked about adjusting the \(\alpha\) level (i.e., instead of determining significance at \(p < .05\), we might adjust and determine a result to be statistically significant if \(p < .005\), depending on how many tests are in our family of tests).
Note what the functions in R do is adjust the \(p\)-value, rather than the \(\alpha\). The Bonferroni method simply multiplies the ‘raw’ p-value by the number of the tests.
In question 4, above, there are 9 tests being performed, but there are 3 in each ‘family’ (each Task).
Try changing your answer to question 4 to use adjust = "none", rather than "bonferroni", and confirm that the p-values are 1/3 of the size.
Šídák
Šídák
- (A bit) more powerful than the Bonferroni method.
- Assumes that all comparisons are independent of one another.
- Less common than Bonferroni method, largely because it is more difficult to calculate (not a problem now we have computers).
The Sidak approach is slightly less conservative than the Bonferroni adjustment.
Doing this with the emmeans package is easy, can you guess how?
Hint: you just have to change the adjust argument in contrast() function.
Tukey
Tukey
- It specifies an exact family significance level for comparing all pairs of treatment means.
- Use Tukey’s method when you are interested in all (or most) pairwise comparisons of means.
As for Šídák, In R we can easily change to Tukey. For instance, if we wanted to conduct pairwise comparisons of the scores of different Diagnosis groups on different Task types (i.e., the interaction):
emm_task <- emmeans(mdl_int, ~ Diagnosis*Task)
contr_task <- contrast(emm_task, method = "pairwise", adjust="tukey")
contr_task## contrast estimate SE df t.ratio
## control grammar - amnesic grammar 20 7.94 36 2.518
## control grammar - huntingtons grammar 40 7.94 36 5.035
## control grammar - control classification 0 7.94 36 0.000
## control grammar - amnesic classification 10 7.94 36 1.259
## control grammar - huntingtons classification 35 7.94 36 4.406
## control grammar - control recognition -15 7.94 36 -1.888
## control grammar - amnesic recognition 15 7.94 36 1.888
## control grammar - huntingtons recognition -15 7.94 36 -1.888
## amnesic grammar - huntingtons grammar 20 7.94 36 2.518
## amnesic grammar - control classification -20 7.94 36 -2.518
## amnesic grammar - amnesic classification -10 7.94 36 -1.259
## amnesic grammar - huntingtons classification 15 7.94 36 1.888
## amnesic grammar - control recognition -35 7.94 36 -4.406
## amnesic grammar - amnesic recognition -5 7.94 36 -0.629
## amnesic grammar - huntingtons recognition -35 7.94 36 -4.406
## huntingtons grammar - control classification -40 7.94 36 -5.035
## huntingtons grammar - amnesic classification -30 7.94 36 -3.776
## huntingtons grammar - huntingtons classification -5 7.94 36 -0.629
## huntingtons grammar - control recognition -55 7.94 36 -6.923
## huntingtons grammar - amnesic recognition -25 7.94 36 -3.147
## huntingtons grammar - huntingtons recognition -55 7.94 36 -6.923
## control classification - amnesic classification 10 7.94 36 1.259
## control classification - huntingtons classification 35 7.94 36 4.406
## control classification - control recognition -15 7.94 36 -1.888
## control classification - amnesic recognition 15 7.94 36 1.888
## control classification - huntingtons recognition -15 7.94 36 -1.888
## amnesic classification - huntingtons classification 25 7.94 36 3.147
## amnesic classification - control recognition -25 7.94 36 -3.147
## amnesic classification - amnesic recognition 5 7.94 36 0.629
## amnesic classification - huntingtons recognition -25 7.94 36 -3.147
## huntingtons classification - control recognition -50 7.94 36 -6.294
## huntingtons classification - amnesic recognition -20 7.94 36 -2.518
## huntingtons classification - huntingtons recognition -50 7.94 36 -6.294
## control recognition - amnesic recognition 30 7.94 36 3.776
## control recognition - huntingtons recognition 0 7.94 36 0.000
## amnesic recognition - huntingtons recognition -30 7.94 36 -3.776
## p.value
## 0.2575
## 0.0004
## 1.0000
## 0.9367
## 0.0026
## 0.6257
## 0.6257
## 0.6257
## 0.2575
## 0.2575
## 0.9367
## 0.6257
## 0.0026
## 0.9993
## 0.0026
## 0.0004
## 0.0149
## 0.9993
## <.0001
## 0.0711
## <.0001
## 0.9367
## 0.0026
## 0.6257
## 0.6257
## 0.6257
## 0.0711
## 0.0711
## 0.9993
## 0.0711
## <.0001
## 0.2575
## <.0001
## 0.0149
## 1.0000
## 0.0149
##
## P value adjustment: tukey method for comparing a family of 9 estimatesWe can also use the following, which doesn’t require the emmeans package. You might see this when you look online for resources. The aov() function is fitting an ANOVA model, and then TukeyHSD() compares between Diagnosis group; between Task type; and between Diagnosis*Task.
Run the code below yourself to see the ouput.
TukeyHSD(aov(Score ~ Diagnosis * Task, data = cog))Scheffe
Scheffe
- It is the most conservative (least powerful) of all tests.
- It controls the family alpha level for testing all possible contrasts.
- It should be used if you have not planned contrasts in advance.
- For testing pairs of treatment means it is too conservative (you should use Bonferroni or Šídák).
emm_task <- emmeans(mdl_int, ~ Diagnosis * Task)
contr_task <- contrast(emm_task, method = "pairwise", adjust="scheffe")
contr_task## contrast estimate SE df t.ratio
## control grammar - amnesic grammar 20 7.94 36 2.518
## control grammar - huntingtons grammar 40 7.94 36 5.035
## control grammar - control classification 0 7.94 36 0.000
## control grammar - amnesic classification 10 7.94 36 1.259
## control grammar - huntingtons classification 35 7.94 36 4.406
## control grammar - control recognition -15 7.94 36 -1.888
## control grammar - amnesic recognition 15 7.94 36 1.888
## control grammar - huntingtons recognition -15 7.94 36 -1.888
## amnesic grammar - huntingtons grammar 20 7.94 36 2.518
## amnesic grammar - control classification -20 7.94 36 -2.518
## amnesic grammar - amnesic classification -10 7.94 36 -1.259
## amnesic grammar - huntingtons classification 15 7.94 36 1.888
## amnesic grammar - control recognition -35 7.94 36 -4.406
## amnesic grammar - amnesic recognition -5 7.94 36 -0.629
## amnesic grammar - huntingtons recognition -35 7.94 36 -4.406
## huntingtons grammar - control classification -40 7.94 36 -5.035
## huntingtons grammar - amnesic classification -30 7.94 36 -3.776
## huntingtons grammar - huntingtons classification -5 7.94 36 -0.629
## huntingtons grammar - control recognition -55 7.94 36 -6.923
## huntingtons grammar - amnesic recognition -25 7.94 36 -3.147
## huntingtons grammar - huntingtons recognition -55 7.94 36 -6.923
## control classification - amnesic classification 10 7.94 36 1.259
## control classification - huntingtons classification 35 7.94 36 4.406
## control classification - control recognition -15 7.94 36 -1.888
## control classification - amnesic recognition 15 7.94 36 1.888
## control classification - huntingtons recognition -15 7.94 36 -1.888
## amnesic classification - huntingtons classification 25 7.94 36 3.147
## amnesic classification - control recognition -25 7.94 36 -3.147
## amnesic classification - amnesic recognition 5 7.94 36 0.629
## amnesic classification - huntingtons recognition -25 7.94 36 -3.147
## huntingtons classification - control recognition -50 7.94 36 -6.294
## huntingtons classification - amnesic recognition -20 7.94 36 -2.518
## huntingtons classification - huntingtons recognition -50 7.94 36 -6.294
## control recognition - amnesic recognition 30 7.94 36 3.776
## control recognition - huntingtons recognition 0 7.94 36 0.000
## amnesic recognition - huntingtons recognition -30 7.94 36 -3.776
## p.value
## 0.6128
## 0.0080
## 1.0000
## 0.9894
## 0.0329
## 0.8852
## 0.8852
## 0.8852
## 0.6128
## 0.6128
## 0.9894
## 0.8852
## 0.0329
## 0.9999
## 0.0329
## 0.0080
## 0.1131
## 0.9999
## 0.0001
## 0.3060
## 0.0001
## 0.9894
## 0.0329
## 0.8852
## 0.8852
## 0.8852
## 0.3060
## 0.3060
## 0.9999
## 0.3060
## 0.0003
## 0.6128
## 0.0003
## 0.1131
## 1.0000
## 0.1131
##
## P value adjustment: scheffe method with rank 8When to use which
For ease of scrolling, we have provided the bulletpoints for each correction below in one place:
Bonferroni
- Use Bonferroni’s method when you are interested in a small number of planned contrasts (or pairwise comparisons).
- Bonferroni’s method is to divide alpha by the number of tests/confidence intervals.
- Assumes that all comparisons are independent of one another.
- It sacrifices slightly more power than Tukey’s method (discussed below), but it can be applied to any set of contrasts or linear combinations (i.e., it is useful in more situations than Tukey).
- It is usually better than Tukey if we want to do a small number of planned comparisons.
Šídák
- (A bit) more powerful than the Bonferroni method.
- Assumes that all comparisons are independent of one another.
- Less common than Bonferroni method, largely because it is more difficult to calculate (not a problem now we have computers).
Tukey
- It specifies an exact family significance level for comparing all pairs of treatment means.
- Use Tukey’s method when you are interested in all (or most) pairwise comparisons of means.
Scheffe
- It is the most conservative (least powerful) of all tests.
- It controls the family alpha level for testing all possible contrasts.
- It should be used if you have not planned contrasts in advance.
- For testing pairs of treatment means it is too conservative (you should use Bonferroni or Šídák).
Lie detecting experiment
Research Questions
Do the Police training materials and the mode of communication (audio vs audiovideo) interact to influence the accuracy of veracity judgements?
Load the data.
Produce a descriptives table for the variables of interest.
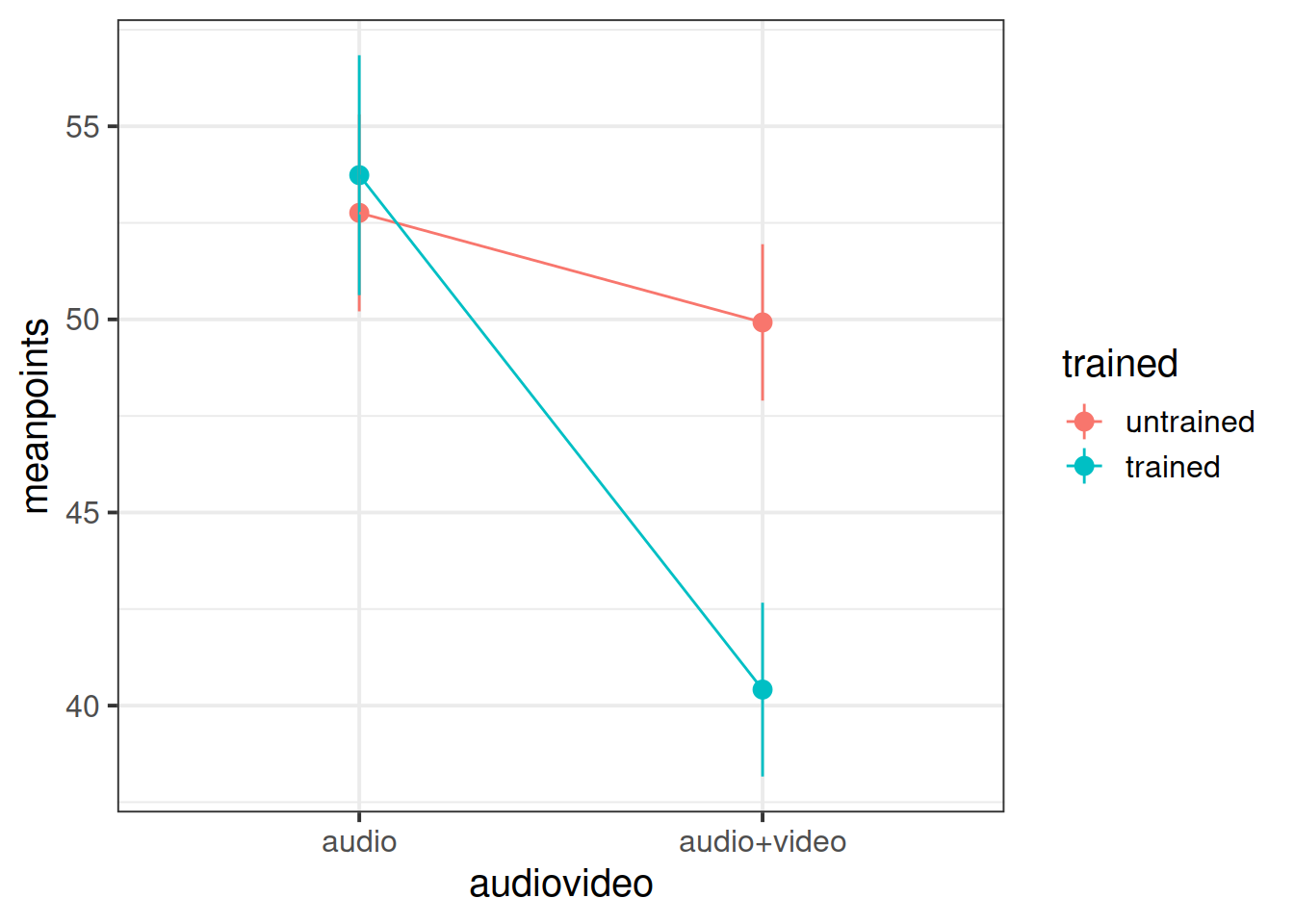
Produce a plot showing the mean points for each condition.
Conduct a two-way ANOVA to investigate research question above.
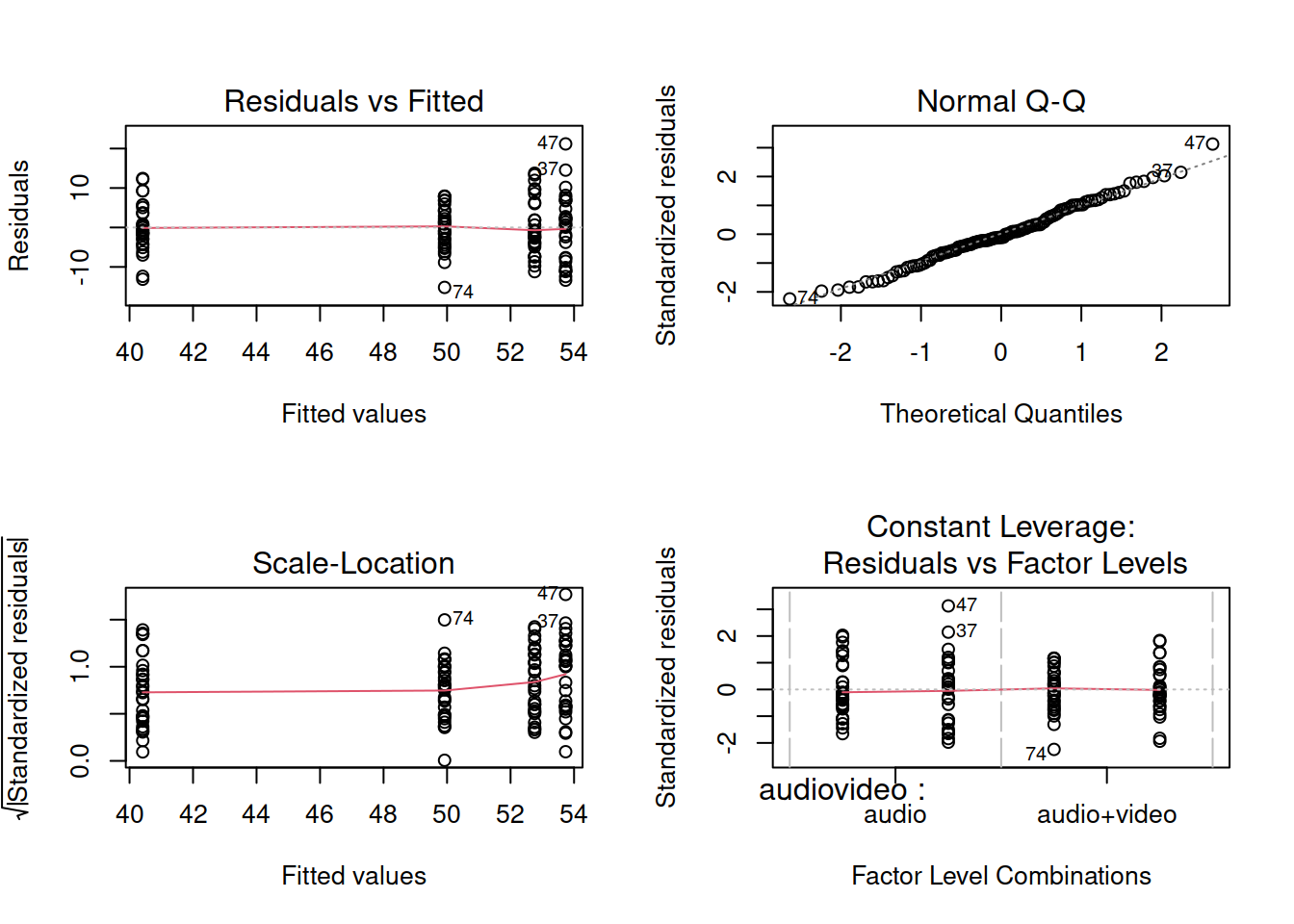
Be sure to check the assumptions!
Write up your results in a paragraph.
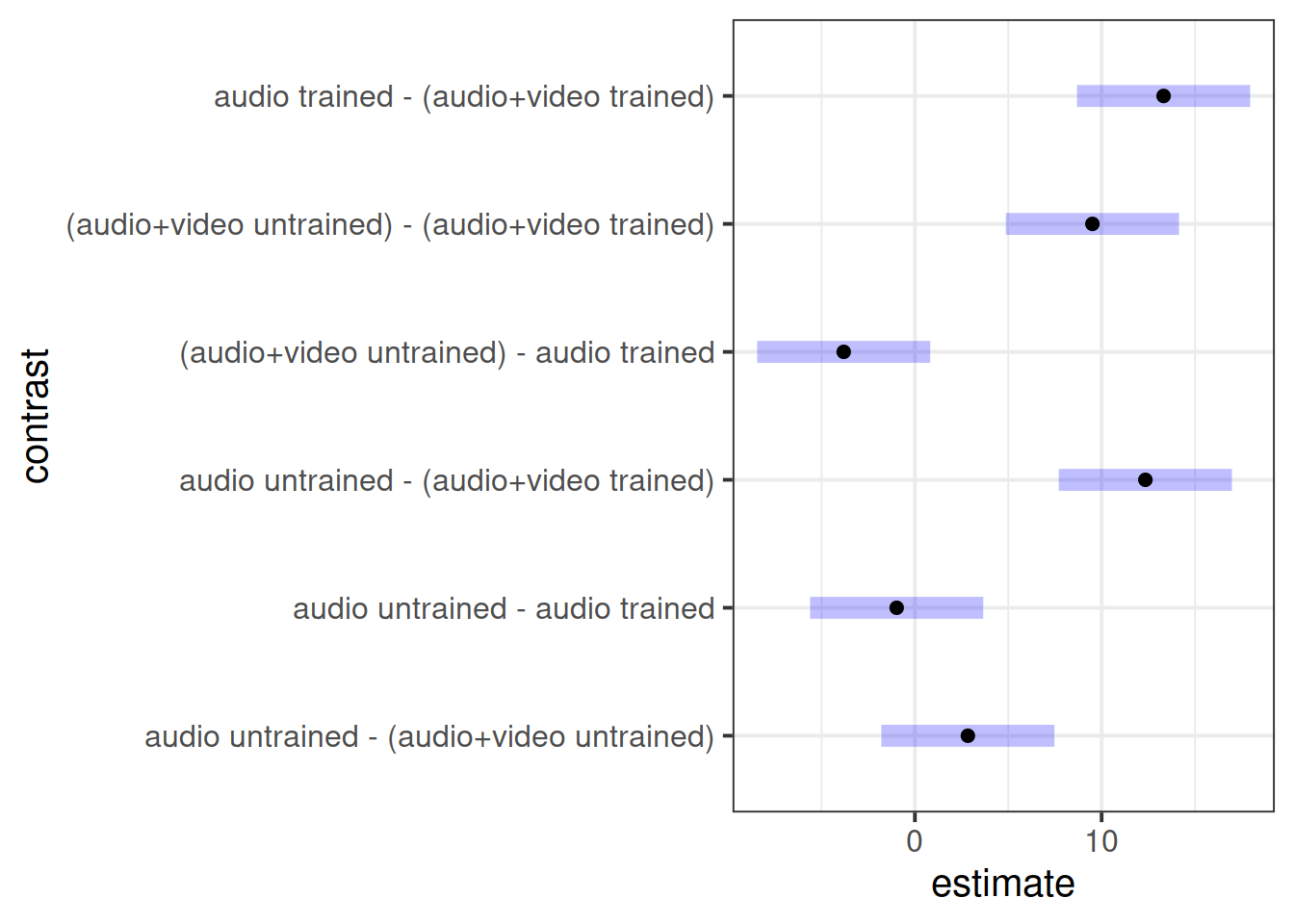
Perform a pairwise comparison of the mean accuracy (as measured by points accrued) across the 2×2 factorial design, making sure to adjust for multiple comparisons by the method of your choice.
Write up your results in a paragraph. Combined with your plot of group means, what do you feel about the Police training materials on using behavioural cues to detect lying?
what defines a ‘family’ of tests is debateable.↩︎