Categorical data
1 A different style of R code
Before we get started on the statistics, we’re going to briefly introduce a crucial bit of R code. We have seen already seen a few examples of R code such as:
And we can actually wrap functions inside functions:
R evaluates code from the inside-out!
You can end up with functions inside functions inside functions …
# Don't worry about what all these functions do,
# it's just an example -
round(mean(log(cumsum(diff(1:10)))))[1] 1We can write in a different style, however, and this may help to keep code tidy and easily readable - we can write sequentially:
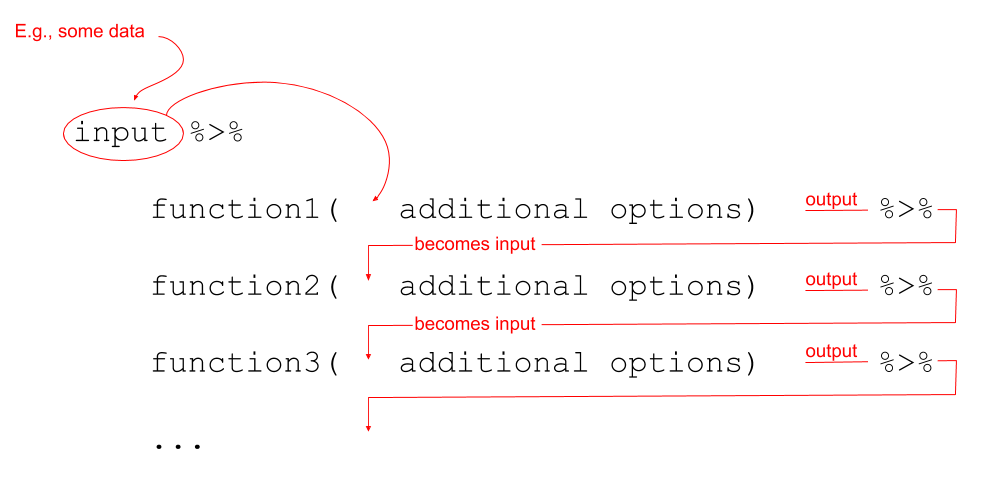
Notice that what we are doing is using a new symbol: %>%
This symbol takes the output of whatever is on it’s left-hand side, and uses it as an input for whatever is on the right-hand side. The %>% symbol gets called a “pipe”.
Let’s see it in action with the starwars2 dataset. The data contains information on various characteristics of characters from Star Wars. Before we can use the pipe operator, %>%, we need to load the tidyverse packages, because that is where %>% is found.
library(tidyverse)
library(patchwork)
starwars2 <- read_csv("https://uoepsy.github.io/data/starwars2.csv")
starwars2 %>%
head()# A tibble: 6 × 6
name height hair_color eye_color homeworld species
<chr> <dbl> <chr> <chr> <chr> <chr>
1 Luke Skywalker 172 blond blue Tatooine Human
2 C-3PO 167 <NA> yellow Tatooine Human
3 R2-D2 96 <NA> red Naboo Droid
4 Darth Vader 202 none yellow Tatooine Human
5 Leia Organa 150 brown brown Alderaan Human
6 Owen Lars 178 brown, grey blue Tatooine Human name height hair_color eye_color
Length:75 Min. : 79.0 Length:75 Length:75
Class :character 1st Qu.:167.5 Class :character Class :character
Mode :character Median :180.0 Mode :character Mode :character
Mean :176.1
3rd Qu.:191.0
Max. :264.0
homeworld species
Length:75 Length:75
Class :character Class :character
Mode :character Mode :character
We can now write code that requires reading it from the inside-out:
or which requires reading it from left to right:
Alderaan Aleen Minor Bespin Bestine IV Cato Neimoidia
3 1 1 1 1
Cerea Champala Chandrila Concord Dawn Corellia
1 1 1 1 2
Coruscant Dathomir Dorin Endor Eriadu
3 1 1 1 1
Geonosis Glee Anselm Haruun Kal Iktotch Iridonia
1 1 1 1 1
Kalee Kamino Kashyyyk Malastare Mirial
1 3 2 1 2
Mon Cala Muunilinst Naboo Nal Hutta Ojom
1 1 8 1 1
Quermia Rodia Ryloth Serenno Shili
1 1 2 1 1
Skako Socorro Springfield Stewjon Sullust
1 1 2 1 1
Tatooine Toydaria Trandosha Troiken Tund
10 1 1 1 1
Utapau Vulpter Zolan
1 1 1 And that long line of code from above:
becomes:
We’re going to use this way of writing a lot throughout the course, and it pairs really well with a group of functions in the tidyverse packages, which were designed to be used in conjunction with %>%.
2 Data Exploration
Once we have collected some data, one of the first things we want to do is explore it - and we can do this through describing (or summarising) and visualising variables.
We are already familiar with the function summary(), which provides high-level information about our data, showing us things such as the minimum and maximum and mean of continuous variables, or the numbers of entries falling into each possible response level for a categorical variable:
summary(starwars2) name height hair_color eye_color
Length:75 Min. : 79.0 Length:75 Length:75
Class :character 1st Qu.:167.5 Class :character Class :character
Mode :character Median :180.0 Mode :character Mode :character
Mean :176.1
3rd Qu.:191.0
Max. :264.0
homeworld species
Length:75 Length:75
Class :character Class :character
Mode :character Mode :character
What we are doing here is providing numeric descriptions of the distributions of values in each variable.
Distribution
The distribution of a variable shows how often different values occur. We’re going to focus on describing and visualising distributions of categorical data.
The graph showing the distribution of a variable shows us where the values are centred, how the values vary, and gives some information about where a typical value might fall. It can also alert you to the presence of outliers (unexpected observations).
3 Unordered Categorical (Nominal) Data
For variables with a discrete number of response options, we can easily measure “how often” values occur in terms of their frequency.
Frequency distribution
A frequency distribution is an overview of all distinct values in some variable and the number of times they occur.
Supposing that we have surveyed the people working in a psychology department and asked them what sub-discipline of psychological research they most strongly identify as working within (If you would like to work along with the reading, the data is available at https://uoepsy.github.io/data/psych_survey.csv).
| Variable Name | Description |
|---|---|
| participant | Subject identifier |
| area | Respondent’s sub-discpline of psychology |
First, we read our data in to R and store it in an object called “psych_disciplines”:
psych_disciplines <- read_csv("https://uoepsy.github.io/data/psych_survey.csv")
psych_disciplines# A tibble: 74 × 2
participant area
<chr> <chr>
1 respondent_1 Differential
2 respondent_2 Social
3 respondent_3 Differential
4 respondent_4 Social
5 respondent_5 Differential
6 respondent_6 Differential
7 respondent_7 Language
8 respondent_8 Language
9 respondent_9 Cognitive Neuroscience
10 respondent_10 Language
# ℹ 64 more rowsWe can get the frequencies of different response levels of the discipline variable by using the following code:
# start with the psych_disciplines dataframe
# %>%
# count() the values in the "area" variable
psych_disciplines %>%
count(area)# A tibble: 5 × 2
area n
<chr> <int>
1 Cognitive Neuroscience 24
2 Developmental 10
3 Differential 20
4 Language 9
5 Social 11In the code above, R knows to look for the area variable inside the psych_disciplines data because we used %>% to “pipe” in the psych_disciplines dataframe.
We could have also done:
# count(data, variable)
count(psych_disciplines, area)# A tibble: 5 × 2
area n
<chr> <int>
1 Cognitive Neuroscience 24
2 Developmental 10
3 Differential 20
4 Language 9
5 Social 11But this would not work:
count(area)Error in group_vars(x) : object ‘area’ not found
Frequency table
To describe a distribution like this, we can simply provide the frequency table.
Let’s store it as an object in R:
# make a new object called "freq_table", and assign it:
# the counts of values of "area" variable in
# the psych_discipline dataframe.
freq_table <-
psych_disciplines %>%
count(area)
# show the object called "freq_table"
freq_table# A tibble: 5 × 2
area n
<chr> <int>
1 Cognitive Neuroscience 24
2 Developmental 10
3 Differential 20
4 Language 9
5 Social 11For a report, we might want to make it a little more easily readable:
Central tendency
Often, we might want to summarise data into a single summary value, reflecting the point at (or around) which most of the values tend to cluster. This is known as a measure of central tendency. For numeric data, we can use measures such as the mean, which you will likely have heard of. For nominal data (unordered categorical data), however, our only option is to use the mode.
Mode
The most frequent value (the value that occurs the greatest number of times).
In our case, the mode is the “Cognitive Neuroscience” category.
Relative frequencies
We might alternatively want to show the percentage of respondents in each category, rather than the raw frequencies.
The percentages show the relative frequency distribution
Relative frequency distribution
A relative frequency distribution shows the proportion of times each value occurs
(contrast this with the frequency distribution which shows the number of times).
Relative frequencies can be written as fractions, percents, or decimals.
In the object “freq_table”, we have a variable called n, which is the frequencies (the number in each category).
The total of this column is equal to the total number of respondents:
# sum all the values in the "n" variable in the "freq_table" object
sum(freq_table$n)[1] 74And therefore, each value in freq_table$n, divided by the total, is equal to the proportion in each category:
( Tip: Proportions are percentages/100. So 0.4 is another way of expressing 40%)
# take the values in the "n" variable from the "freq_table" object,
# and divide by the sum of all the values in the "n" variable in "freq_table"
freq_table$n/sum(freq_table$n)[1] 0.3243243 0.1351351 0.2702703 0.1216216 0.1486486We can then simply add the proportions as a new column to our table of frequencies by assigning the values we just calculated to a new variable:
# the variable "prop" in the "freq_table" object is now assigned
# the values we calculated above (the proportions)
freq_table$prop <- freq_table$n/sum(freq_table$n)
# print the "freq_table" object
freq_table# A tibble: 5 × 3
area n prop
<chr> <int> <dbl>
1 Cognitive Neuroscience 24 0.324
2 Developmental 10 0.135
3 Differential 20 0.270
4 Language 9 0.122
5 Social 11 0.149
However, we can also do this within a sequence of pipes (%>%). To do so, we use a new function called mutate().
The mutate() function is used to add or modify variables to data.
Note: Inside mutate(), we don’t have to keep using the dollar sign $, as we have already told it what data to look for variables in.
To ensure that our additions/modifications of variables are stored in R’s environment (rather than simply printed out), we need to reassign the name of our dataframe:
We can actually add this step to our earlier code:
# make a new object called "freq_table", and assign it:
# the counts of values of "area" variable in
# the psych_discipline dataframe.
# from there, 'mutate' such that there is a variable called "prop" which
# has the values of the "n" variable divided by the sum of the "n" variable.
freq_table <-
psych_disciplines %>%
count(area) %>%
mutate(
prop = n/sum(n)
)
# show the object called "freq_table"
freq_table# A tibble: 5 × 3
area n prop
<chr> <int> <dbl>
1 Cognitive Neuroscience 24 0.324
2 Developmental 10 0.135
3 Differential 20 0.270
4 Language 9 0.122
5 Social 11 0.149Visualising
“By visualizing information, we turn it into a landscape that you can explore with your eyes. A sort of information map. And when you’re lost in information, an information map is kind of useful.”_ David McCandless
We’re going to now make our first steps into the world of data visualisation. R is an incredibly capable language for creating visualisations of almost any kind. It is used by many media companies (e.g., the BBC), and has the capability of producing 3d visualisations, animations, interactive graphs, and more.
We are going to use the most popular R package for visualisation, ggplot2. This is actually part of the tidyverse, so if we have an Rmarkdown document and have loaded the tidyverse packages at the start (by using library(tidyverse)), then ggplot2 will be loaded too).
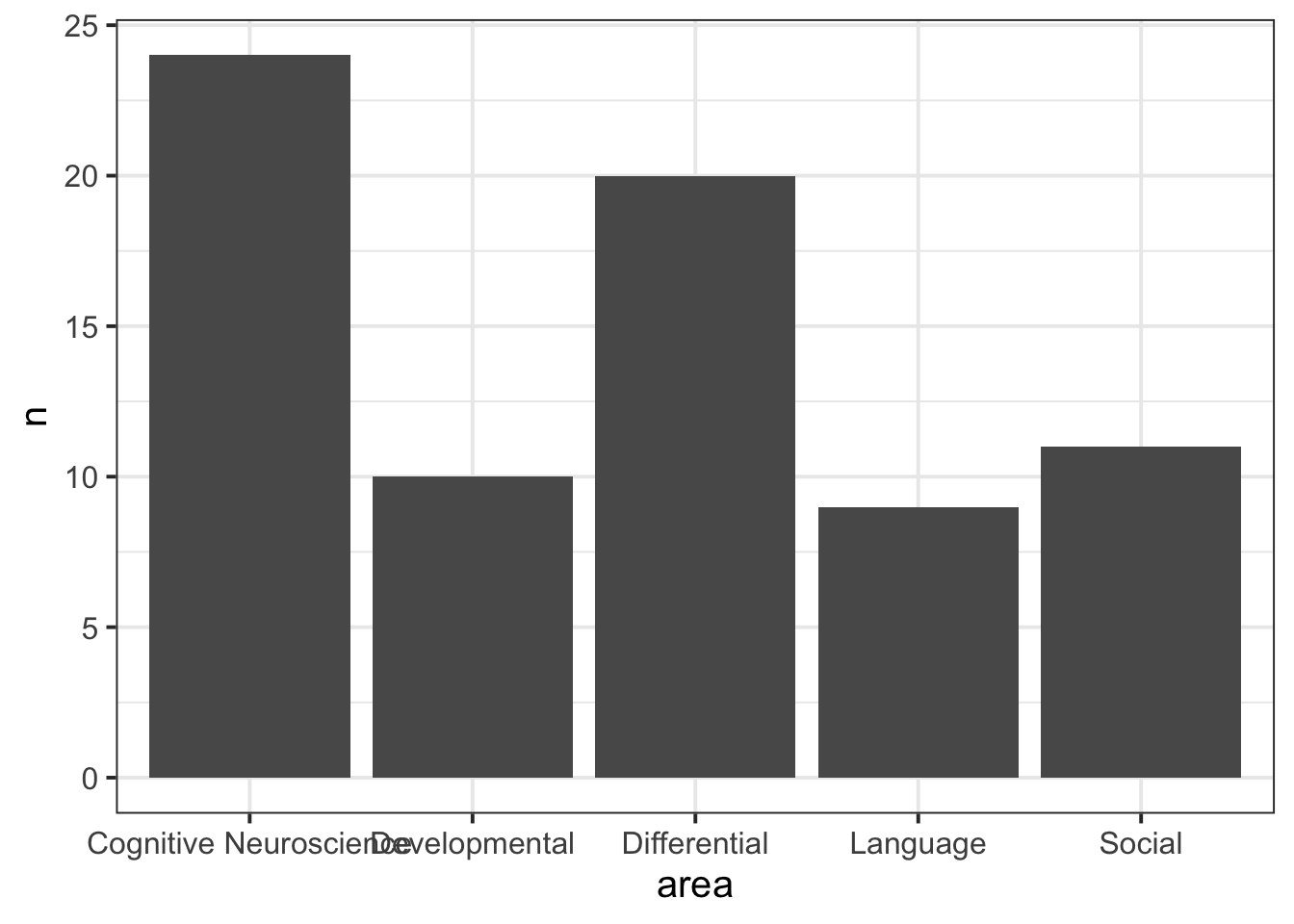
Recall our frequency distribution table:
# show the object called "freq_table"
freq_table# A tibble: 5 × 3
area n prop
<chr> <int> <dbl>
1 Cognitive Neuroscience 24 0.324
2 Developmental 10 0.135
3 Differential 20 0.270
4 Language 9 0.122
5 Social 11 0.149We can plot these values as a bar chart:

ggplot components
Note the key components of the ggplot code.
-
data =where we provide the name of the dataframe. -
aes =where we provide the aesthetics. These are things which we map from the data to the graph. For instance, the x-axis, or if we wanted to colour the columns/bars according to some aspect of the data.
Then we add (using +) some geometry. These are the shapes (in our case, the columns/bars), which will be put in the correct place according to what we specified in aes().
-
+ geom_col()Adds columns to the plot.
Use these as reference for when you want to make changes to the plots you create.
Additionall, remember that google is your friend - there are endless forums with people asking how to do something in ggplot, and you can just copy and paste bits of code to add to your plots!
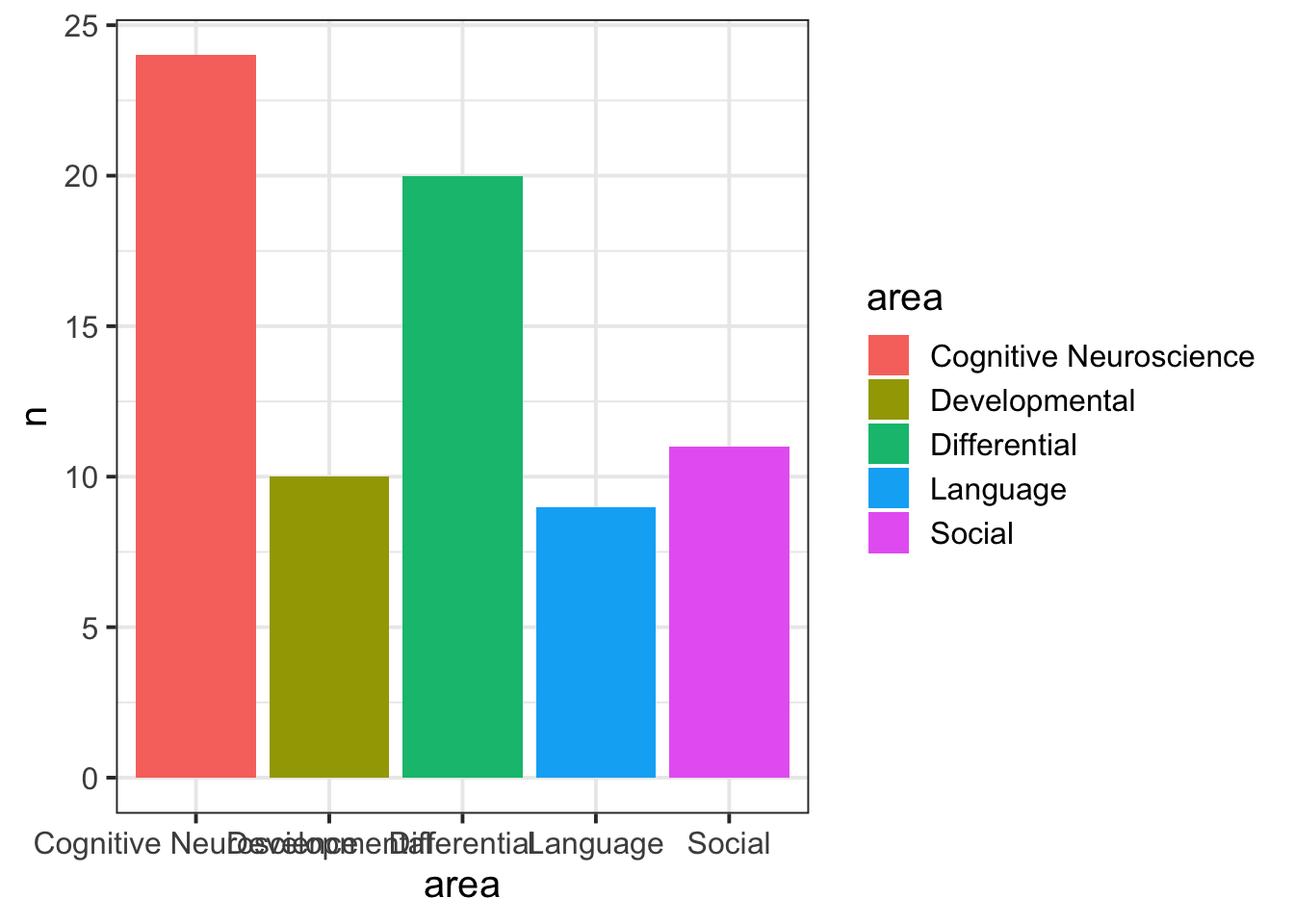
- Fill the geoms:
- Change the axis labels:
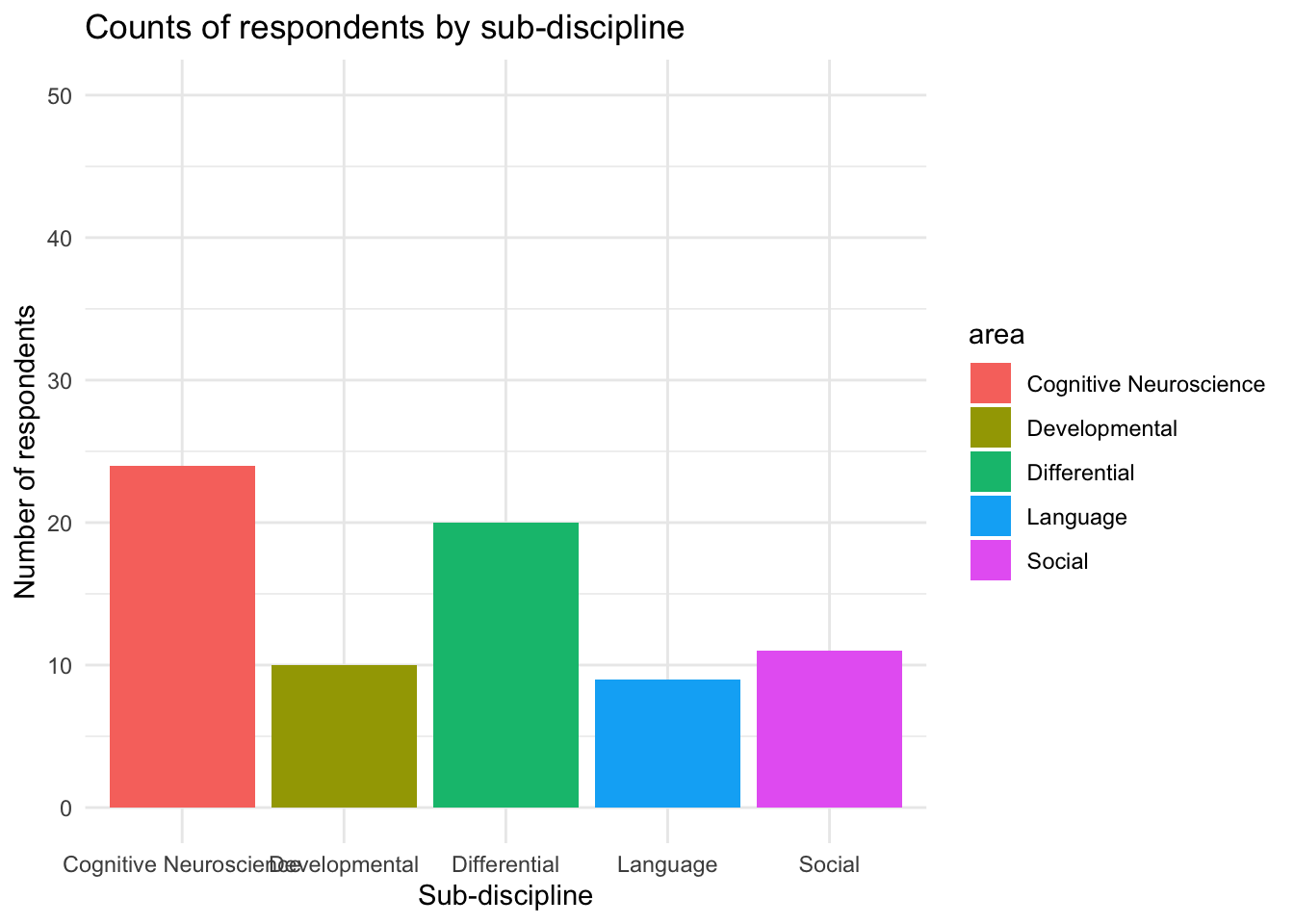
ggplot(data = freq_table, aes(x = area, y = n, fill = area)) +
geom_col()+
labs(title="Counts of respondents by sub-discipline", y = "Number of respondents", x = "Sub-discipline")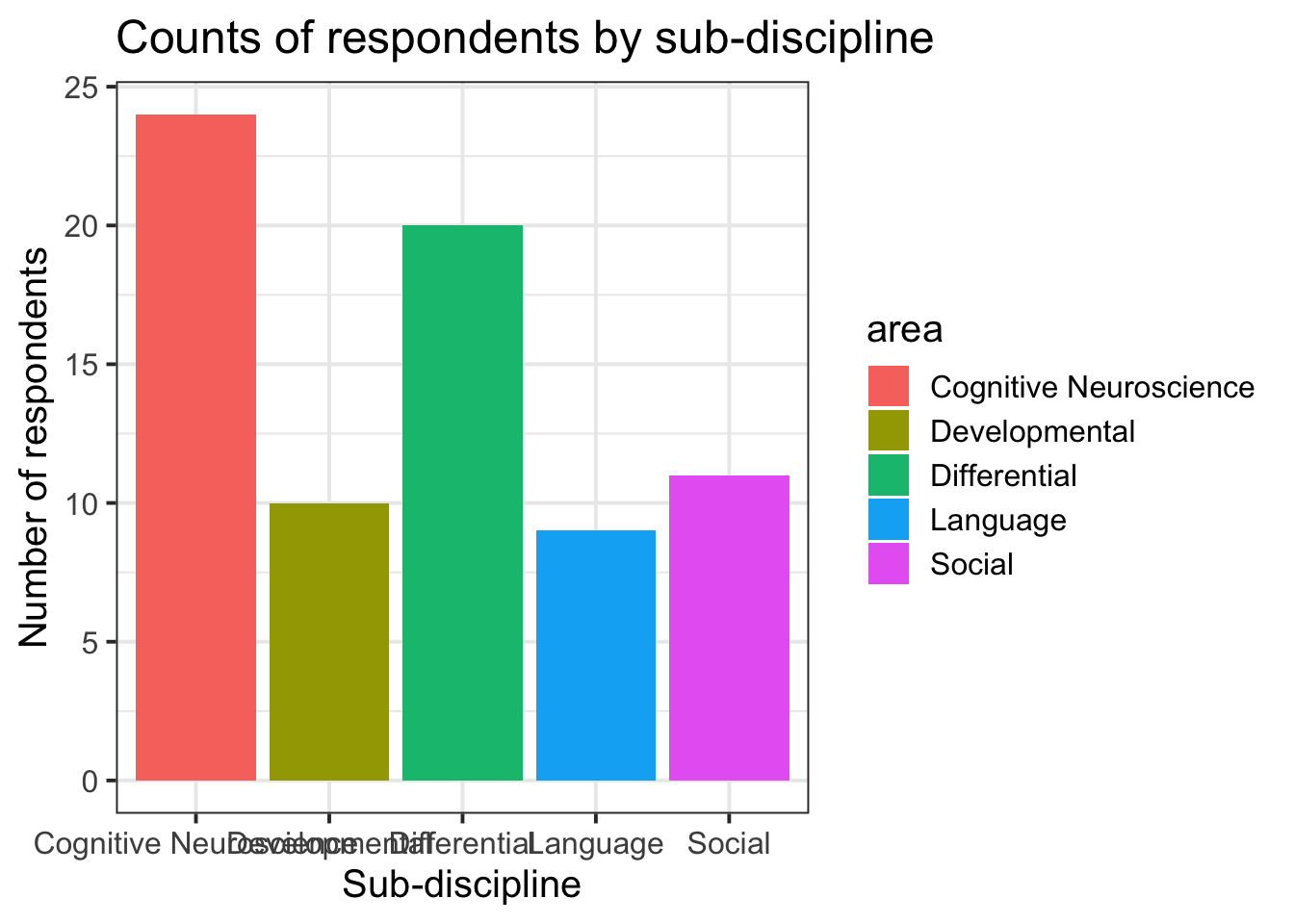
- Change the geom:
(Note that usinggeom_colhad the y axis starting at 0, but geom_point starts just below the lowest value.
# note that we also need to change "fill = area" to "col = area".
ggplot(data = freq_table, aes(x = area, y = n, col = area)) +
geom_point()+
labs(title="Counts of respondents by sub-discipline", y = "Number of respondents", x = "Sub-discipline")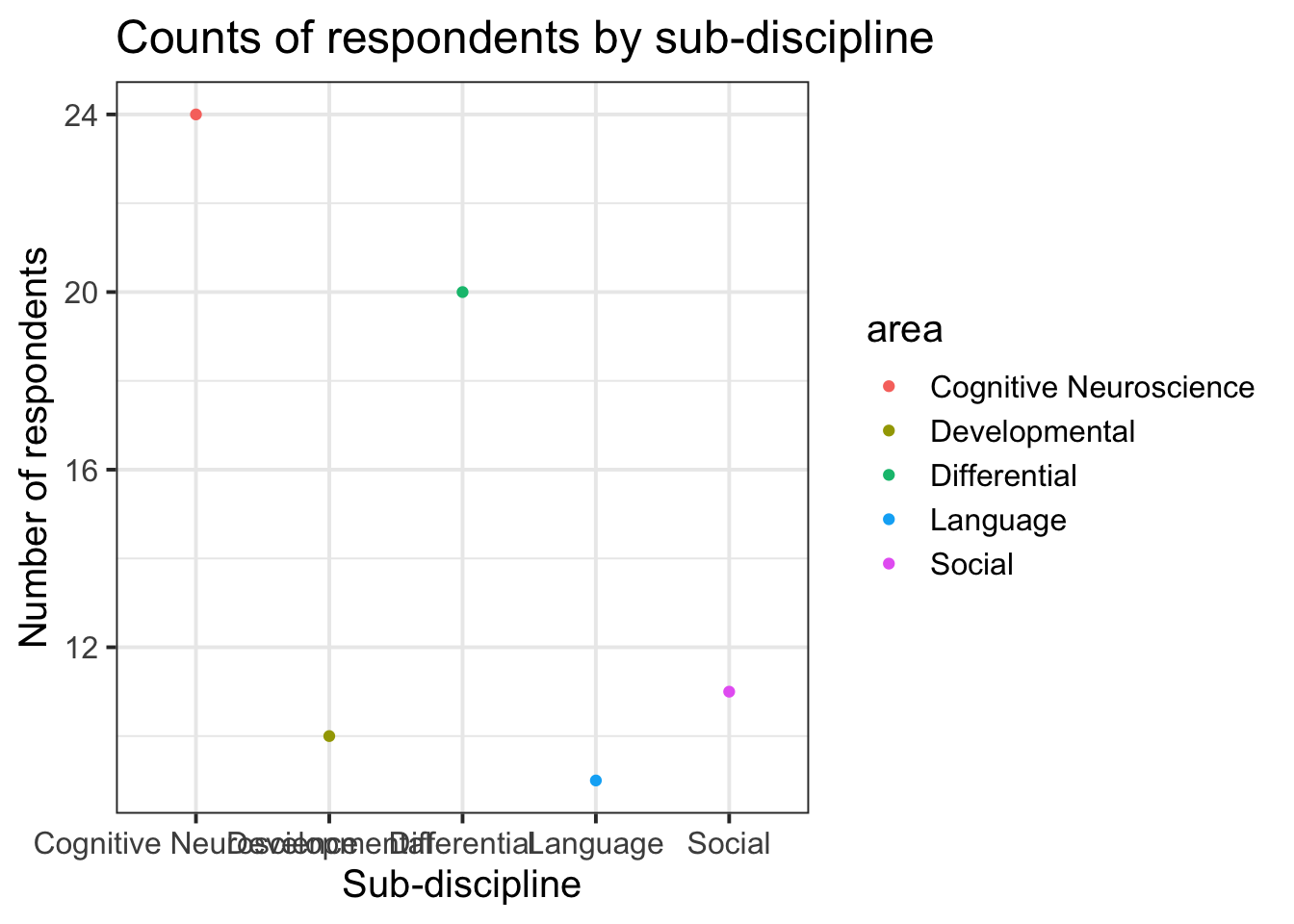
- Change the limits of the axes:
ggplot(data = freq_table, aes(x = area, y = n, fill = area)) +
geom_col()+
labs(title="Counts of respondents by sub-discipline", y = "Number of respondents", x = "Sub-discipline")+
ylim(0,50)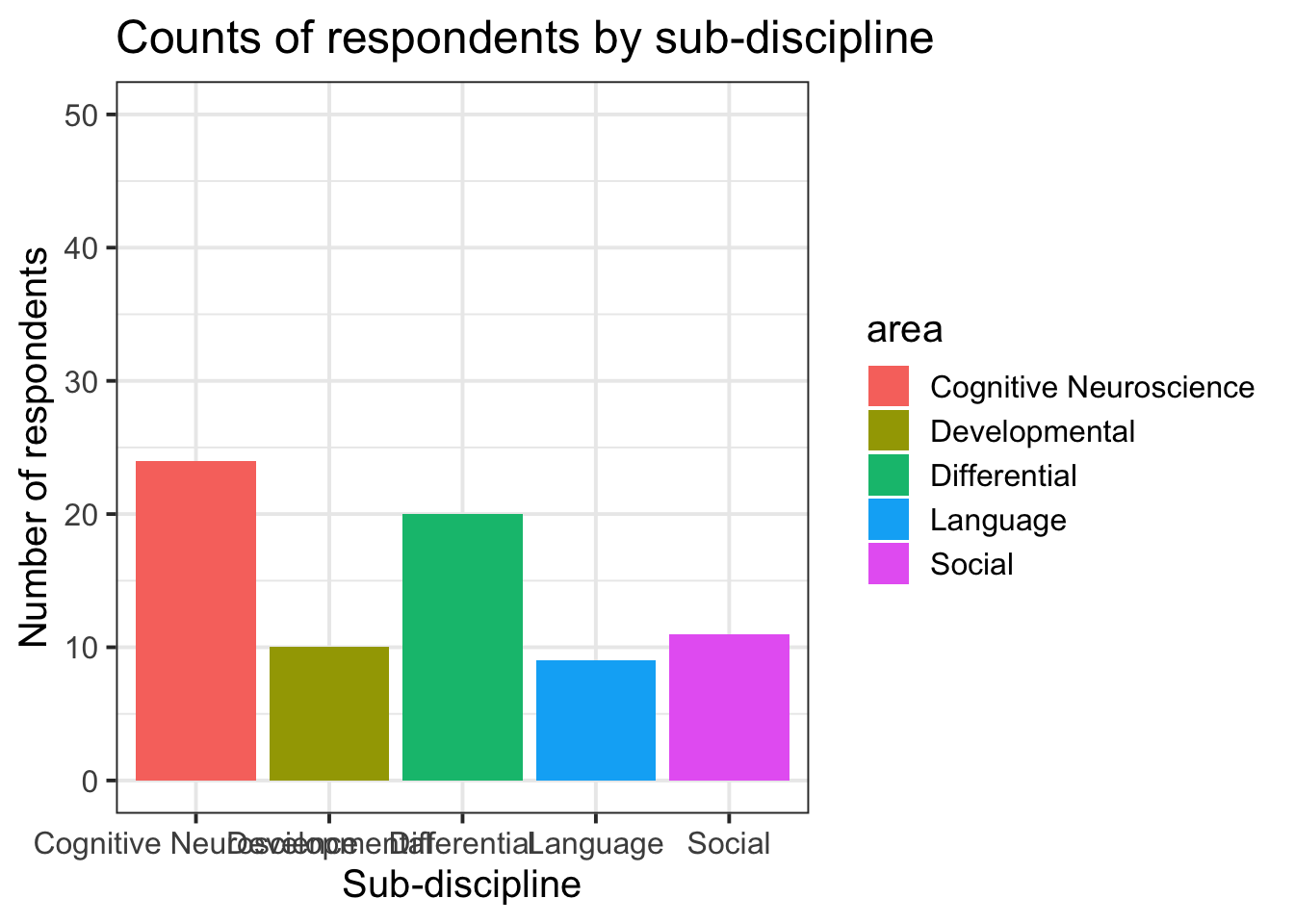
- Remove (or reposition) the legend:
# setting legend.position as "bottom" would put it at the bottom!
ggplot(data = freq_table, aes(x = area, y = n, fill = area)) +
geom_col()+
labs(title="Counts of respondents by sub-discipline", y = "Number of respondents", x = "Sub-discipline")+
ylim(0,50)+
theme(legend.position = "none") 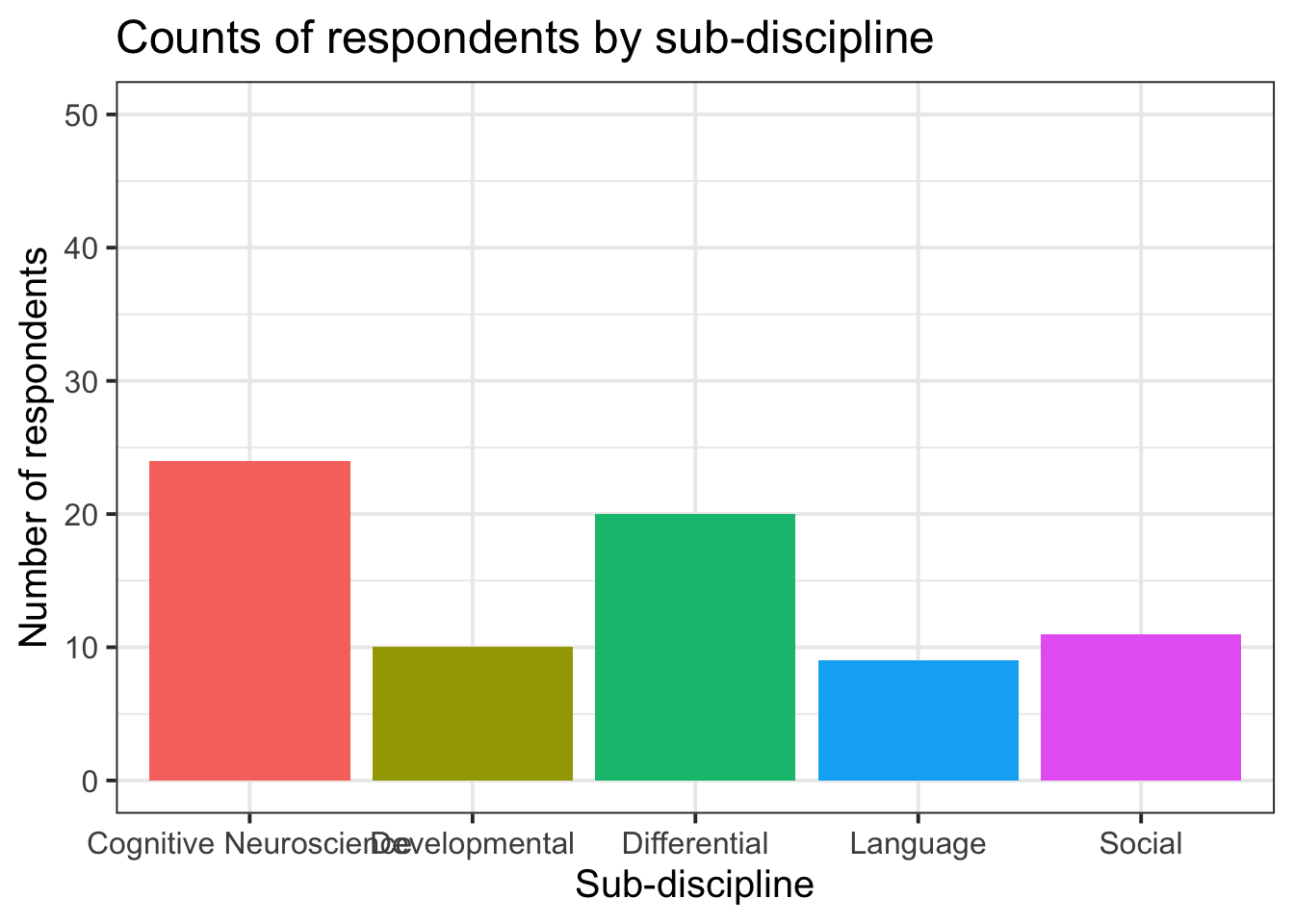
- Changing the theme:
4 Ordered Categorical (Ordinal) Data
Recall that ordinal data is categorical data which has a natural ordering of the possible responses. One of the most common examples of ordinal data which you will encounter in psychology is the Likert Scale. You will probably have come across these before, perhaps when completing online surveys or questionnaires.
Likert Scale
A five or seven point scale on which an individual express how much they agree or disagree with a particular statement.
With Likert data, there is a set of discrete response options (it is categorical data). The response options can be ranked, making it ordered categorical ( strongly disagree < disagree < neither < agree < strongly agree ). Importantly, the distance between responses is not measurable.
Frequency table
Let’s suppose that as well as collecting information on the sub-discipline of psychology they identified with, we also asked our respondents to rate their level of happiness from 1 to 5, as well as their job satisfaction from 1 to 5.
| Variable Name | Description |
|---|---|
| participant | Subject identifier |
| happiness | Respondent’s level of happiness from 1 to 5 |
| job_sat | Respondent’s level of job satisfaction from 1 to 5 |
psych_survey <- read_csv("https://uoepsy.github.io/data/psych_survey2.csv")
psych_survey# A tibble: 74 × 3
participant happiness job_sat
<chr> <dbl> <dbl>
1 respondent_1 3 3
2 respondent_2 3 4
3 respondent_3 2 5
4 respondent_4 4 5
5 respondent_5 3 5
6 respondent_6 4 4
7 respondent_7 4 2
8 respondent_8 5 5
9 respondent_9 1 5
10 respondent_10 3 4
# ℹ 64 more rowsFor these questions (variables happiness and job_sat), we could do the same thing as we did above for unordered categorical data, and summarise this into frequencies:
# take the "psych_survey" dataframe %>%
# count() the values in the "happiness" variable
psych_survey %>%
count(happiness)# A tibble: 5 × 2
happiness n
<dbl> <int>
1 1 6
2 2 13
3 3 27
4 4 21
5 5 7# take the "psych_survey" dataframe %>%
# count() the values in the "job_sat" variable
psych_survey %>%
count(job_sat)# A tibble: 5 × 2
job_sat n
<dbl> <int>
1 1 3
2 2 6
3 3 11
4 4 16
5 5 38Central tendency
We could again use the Mode - the most common value - to summarise this data. However, because the responses are ordered, it can be more useful to think about the percentages of respondents in and below/above each category. For instance, we might want to talk about asking which category has 50% of the observations in a lower category, and 50% of the observations in a higher category. This is mid-point is known as the Median.
Median
The value for which 50% of observations a lower and 50% are higher. It is the mid-point of a list of ordered values.
To find the median:
- rank order the values
- find the middle value:
- If there are \(n\) values, find the value at position \(\frac{n+1}{2}\).
- If \(n\) is even, \(\frac{n+1}{2}\) will not be a whole number.
For instance, if \(n = 20\), you are looking for the \(\frac{n+1}{2} = \frac{20+1}{2} = 10.5^{th}\) value.- When calculating the median for ordinal data, if the \(\frac{n}{2}^{th}\) and \(\frac{n+1}{2}^{th}\) values are different, report both.
- When calculating the median for numeric data, report the midpoint of the \(\frac{n}{2}^{th}\) and \(\frac{n+1}{2}^{th}\) values.
- If there are \(n\) values, find the value at position \(\frac{n+1}{2}\).
You can tell R explicitly that a variable is of a certain type using functions such as as.factor(), as.numeric(), and so on.
You may notice that we haven’t done this yet with the data we have been working with in so far today:
# inside the "psych_survey" dataframe, take ($) the "happiness" variable,
# and tell me what type/class it is
class(psych_survey$happiness)[1] "numeric"This is because there are some benefits to letting R think your data is numeric, even when it is not. It means we can use functions such as median() to quickly find the median:
# inside the "psych_survey" dataframe, take ($) the "happiness" variable,
# and find the median
median(psych_survey$happiness)[1] 3While we can make R treat this data is numeric, it is important to remember that it is actually measured on an ordinal scale.
For example, if the median falls between levels, R will tell us that the median is the mid-point:
But because our data is ordinal, then we know that 2.5 is not a valid response.
We can also use functions such as min() and max() to find the minimum and maximum values:
Cumulative percentages, Quartiles
In calculating the median, we are going beyond talking about the relative frequencies (i.e., the percentage in each category), to talking about the cumulative percentage.
Cumulative percentage
Cumulative percentages are another way of expressing a frequency distribution.
They are the successive addition of percentages in each category. For example, the cumulative percentage for the 3rd category is the percentage of respondents in the 1st, 2nd and 3rd category:
| Category | Frequency count (n) | Relative frequency (%) | Cumulative frequency | Cumulative percentage |
|---|---|---|---|---|
| Response 1 | 10 | 13.33 | 10 | 13.33 |
| Response 2 | 10 | 13.33 | 20 | 26.67 |
| Response 3 | 20 | 26.67 | 40 | 53.33 |
| Response 4 | 25 | 33.33 | 65 | 86.67 |
| Response 5 | 10 | 13.33 | 75 | 100.00 |
We saw before how we can calculate the proportions/percentages in each category:
( Note: We multiply by 100 here to turn the proportion into a percentage)
# take the "psych_survey" dataframe %>%
# count() the values in the "happiness" variable (creates an "n" column), and
# from there, 'mutate' such that there is a variable called "percent" which
# has the values of the "n" variable divided by the sum of the "n" variable.
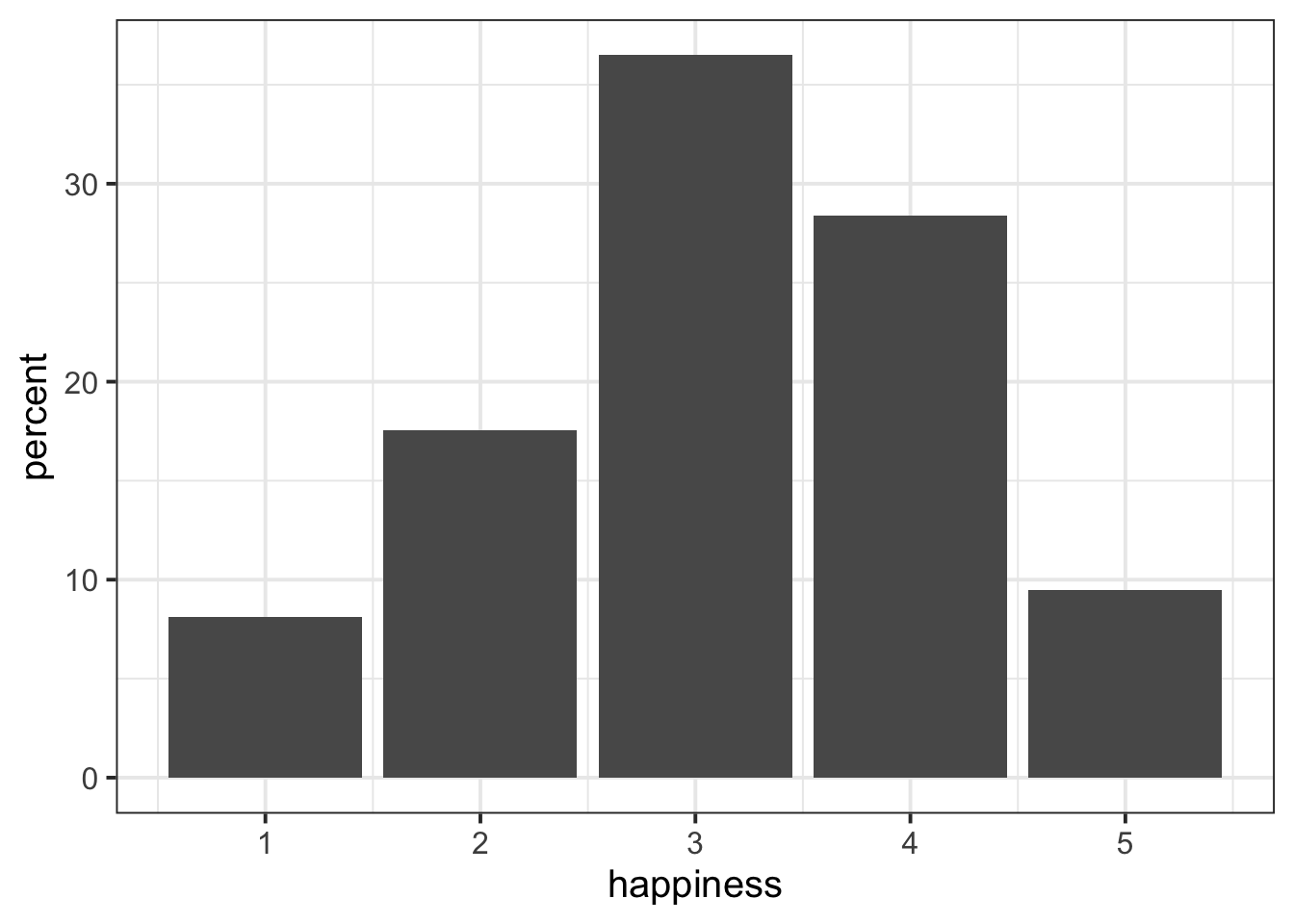
psych_survey %>%
count(happiness) %>%
mutate(
percent = n/sum(n)*100
)# A tibble: 5 × 3
happiness n percent
<dbl> <int> <dbl>
1 1 6 8.11
2 2 13 17.6
3 3 27 36.5
4 4 21 28.4
5 5 7 9.46We can add another variable, and make it the cumulative percentage, by using the cumsum() function.
# take the "psych_survey" dataframe %>%
# count() the values in the "happiness" variable (creates an "n" column), and
# from there, 'mutate' such that there is a variable called "percent" which
# has the values of the "n" variable divided by the sum of the "n" variable,
# and also make a variable called "cumulative_percent" which is the
# successive addition of the values in the "percent" variable
psych_survey %>%
count(happiness) %>%
mutate(
percent = n/sum(n)*100,
cumulative_percent = cumsum(percent)
)# A tibble: 5 × 4
happiness n percent cumulative_percent
<dbl> <int> <dbl> <dbl>
1 1 6 8.11 8.11
2 2 13 17.6 25.7
3 3 27 36.5 62.2
4 4 21 28.4 90.5
5 5 7 9.46 100 Think about why this will not work:
Error: object ‘percent’ not found
Answer: Inside the mutate() function, we are trying to assign the cumulative_percent variable based on the values of the percent variable. But in the code above, percent gets defined after cumulative_percent, and so it will not work. Hence the error message (“percent not found”).
While the median splits the data in two (50% either side), you will often see data being split into four equal blocks.
The points which divide the four blocks are known as quartiles.
Quartiles
Quartiles are the points in rank-ordered data below which falls 25%, 50%, and 75% of the data.
- The first quartile is the first category for which the cumulative percentage is \(\geq 25\%\).
- The median is the first category for which the cumulative percentage is \(\geq 50\%\).
- The third quartile is the first category for which the cumulative percentage is \(\geq 75\%\).
By looking at the quartiles, it gives us an idea of how spread out the data is.
As an example, if we had 10 categories A, B, C, D, E, F, G, H, I, J, and we knew that:
- \(Q_1\) (the \(1^{st}\) quartile) = G,
- \(Q_2\) (the \(2^{nd}\) quartile, the median) = H,
- \(Q_3\) (the \(3^{rd}\) quartile) = H,
This tells us that the first 25% of the data falls in one of the categories from A to G (quite a large range), the second 25% falls in categories G and H (a small range), and the third 25% of the data falls entirely in category H.
So a lot of the data is between G and H, with the data being more sparse in the lower and higher categories.
Looking ahead to numeric data
We will talk about quartiles in numeric data too, where we commonly use the difference between the first and third quartile as a measure of how spread out the data are. This gets known as the inter-quartile range (IQR).
Visualising
We can visualise ordered categorical data in the same way we did for unordered.
First we save our frequencies/percentages as a new object:
Then we give that object to our ggplot code, with the appropriate aes() mappings:
5 Glossary
- distribution: How often different possible values in a variable occur.
- frequency: Number of occurrences (count) in a given response value.
- relative frequency: Percentage/proportion of occurrences in a given response value.
- cumulative percentage: Percentage of occurrences in or below a given reponse value (requires ordered data).
- mode: Most common value.
-
median: Middle value.
-
%>%Takes the output of whatever is on the LHS and gives it as the input of whatever is on the RHS. -
count()Counts the number of occurrences of each unique value in a variable. -
mutate()Used to add variables to the dataframe, or modify existing variables. -
min()Returns the minimum value of a variable. -
max()Returns the maximum value of a variable. -
median()Returns the median value of a variable. -
ggplot()Creates a plot. Takesdata=and a set of mappingsaes()from the data to properties of the plot (e.g., x/y axes, colours). -
geom_col()Adds columns to a ggplot.