More SEM
In this final week we are going to address some practical issues that commonly arise in Structural Equation Modeling: Non-normality, ordinal-categorical endogeneous variables, and missingness. After that, we’re going to look briefly into how to simulate data based on a structural equation model.
Practical issues in SEM
One of the key assumptions of the models we have been fitting so far is that our variables are all normally distributed, and are continuous. Why?
Recall how we first introduced the idea of estimating a SEM: comparing the observed covariance matrix with our model implied covariance matrix. This heavily relies on the assumption that our observed covariance matrix provides a good representation of our data. Unfortunately, this is true if we have a multivariate normal distribution, but becomes more difficult when our variables are either not continuous or not normally distributed.
MVN
The multivariate normal distribution is fundamentally just the extension of our univariate normal (the bell-shaped curve we are used to seeing) to more dimensions. The condition which needs to be met is that every linear combination of the \(k\) variables has a univariate normal distribution. It is denoted by \(N \sim(\mathbf{\mu}, \mathbf{\Sigma})\) (note that the bold font indicates that \(\mathbf{\mu}\) and \(\mathbf{\Sigma}\) are, respectively, a vector of means and a matrix of covariances).
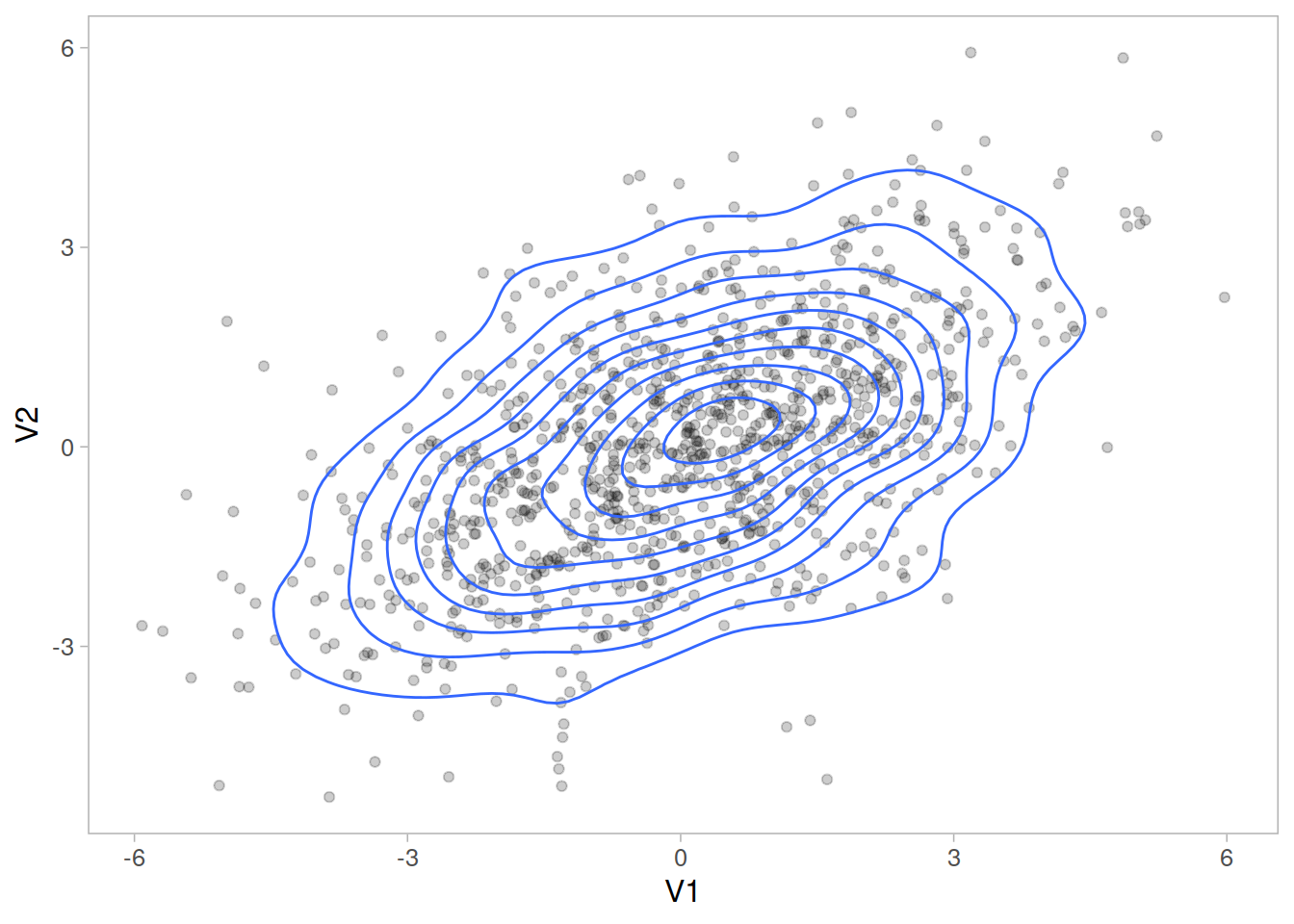
The idea of the multivariate normal can be a bit difficult to get to grips with in part because there’s not an intuitive way to visualise it once we move beyond 2 or 3 variables.
Figure 1: bivariate normal
You may also see this represented by a contour plot, which should look similar to the above 3D plot viewed from above.
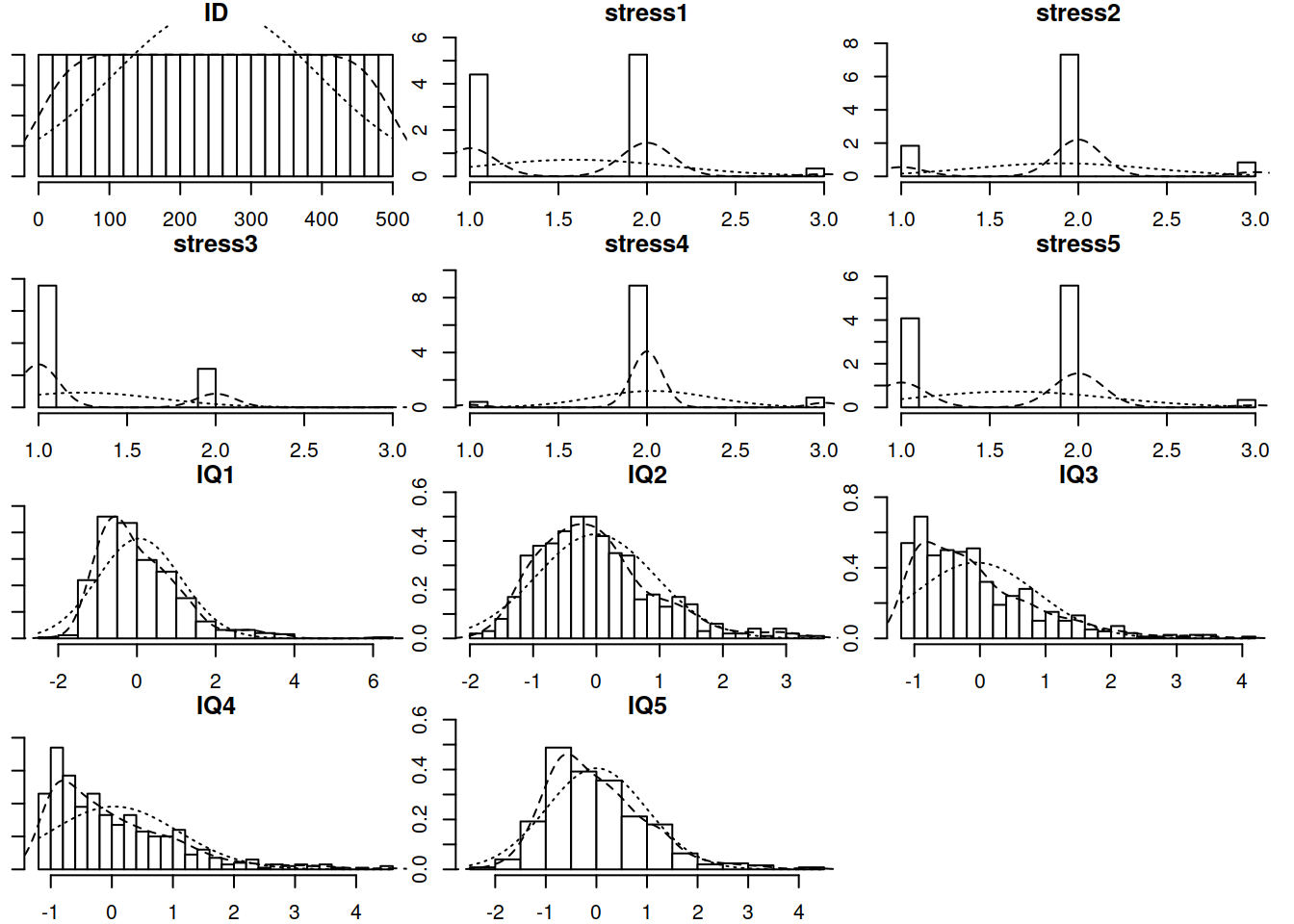
In the first example for this week a researcher is interested in the effects of prenatal stress on child cognitive outcomes. She has a 5-item measure of prenatal stress and a 5 subtest measure of child cognitive ability, collected for 500 mother-infant dyads.
- The data is available as a .csv file here: https://uoepsy.github.io/data/stressIQ.csv
Read in the data and explore it.
You may remember the pairs.panels() function from the psych package. Another useful one from the same package is multi.hist().
Ordered-Categorical Endogenous Variables
When can we treat ordinal data as if it is continuous?
This can be a contentious issue for some statisticians, as it is arguable that ordinal data is not continuous, so we should not treat it as such. Much research using SEM centers around questionnaire data, which lends itself to likert data (for instance, “strongly agree,”“agree,”“neither agree nor disagree,”“disagree,”“strongly disagree”). An often used rule of thumb, is that likert data with \(\geq 5\) levels can be treated as if they are continuous without unduly influencing results (see Johnson, D.R., & Creech, J.C. (1983). Ordinal measures in multiple indicator models: A simulation study of categorization error).
What to do
In R, lavaan will automatically switch to a categorical estimator if we tell it that we have some ordered-categorical variables. We can use the ordered = c("item1name","item2name","item3name", ...) argument.
This is true for both the cfa() and sem() functions.
The prenatal stress questionnaire items are measured on a 3-point scale.
Read in the data and fit a one-factor confirmatory factor analysis specifying any ordered-categorical variables in order to use an appropriate estimation method.
Inspect your model output. Notice that you have an extra column - we now have ‘robust’ values for the fit statistics (those that appear in the right-hand column under ‘robust’) to check that our model fits well.
Another new thing we have is the presence of ‘thresholds’ in our output. These are two thresholds per item in this example because we have a three-point response scale. Estimation of ordered-categorical variables involves constructing a hypothetical continuum underlying our categories, with “thresholds” being the points on this continuum where an observation moves from being one category to the next.
Non-normality
What counts as “non-normal?”
There are various measures of both skewness and kurtosis (see Joanes, D.N. and Gill, C.A (1998). Comparing measures of sample skewness and kurtosis if you’re interested). In addition, there are various suggested rules of thumb we can follow. Below are the most common:
Skewness rules of thumb:
- \(|skew| < 0.5\): fairly symmetrical
- \(0.5 < |skew| < 1\): moderately skewed
- \(1 < |skew|\): highly skewed
Kurtosis rule of thumb:
- \(Kurtosis > 3\): Heavier tails than the normal distribution. Possibly problematic
What to do
When faced with variables which appear to deviate from normality, we should ideally use a robust estimator that corrects for any bias in the standard errors induced by non-normality, while also providing a corrected \(\chi^2\) statistic to more accurately capture the misfit of our model.
There are a few robust estimators in lavaan, but one of the more frequently used ones is “MLR” (maximum likelihood with robust SEs).
You can find all the other options at https://lavaan.ugent.be/tutorial/est.html.
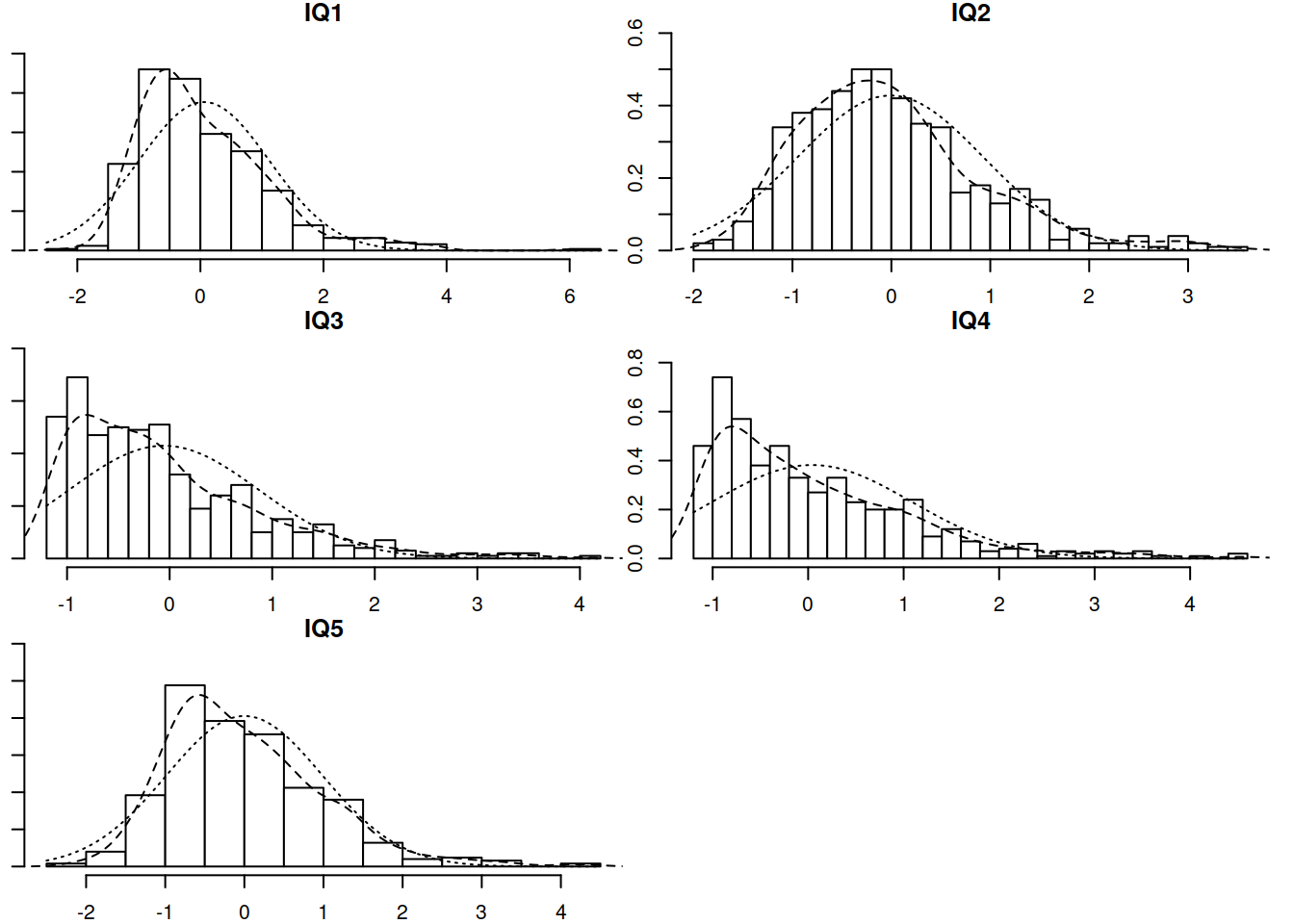
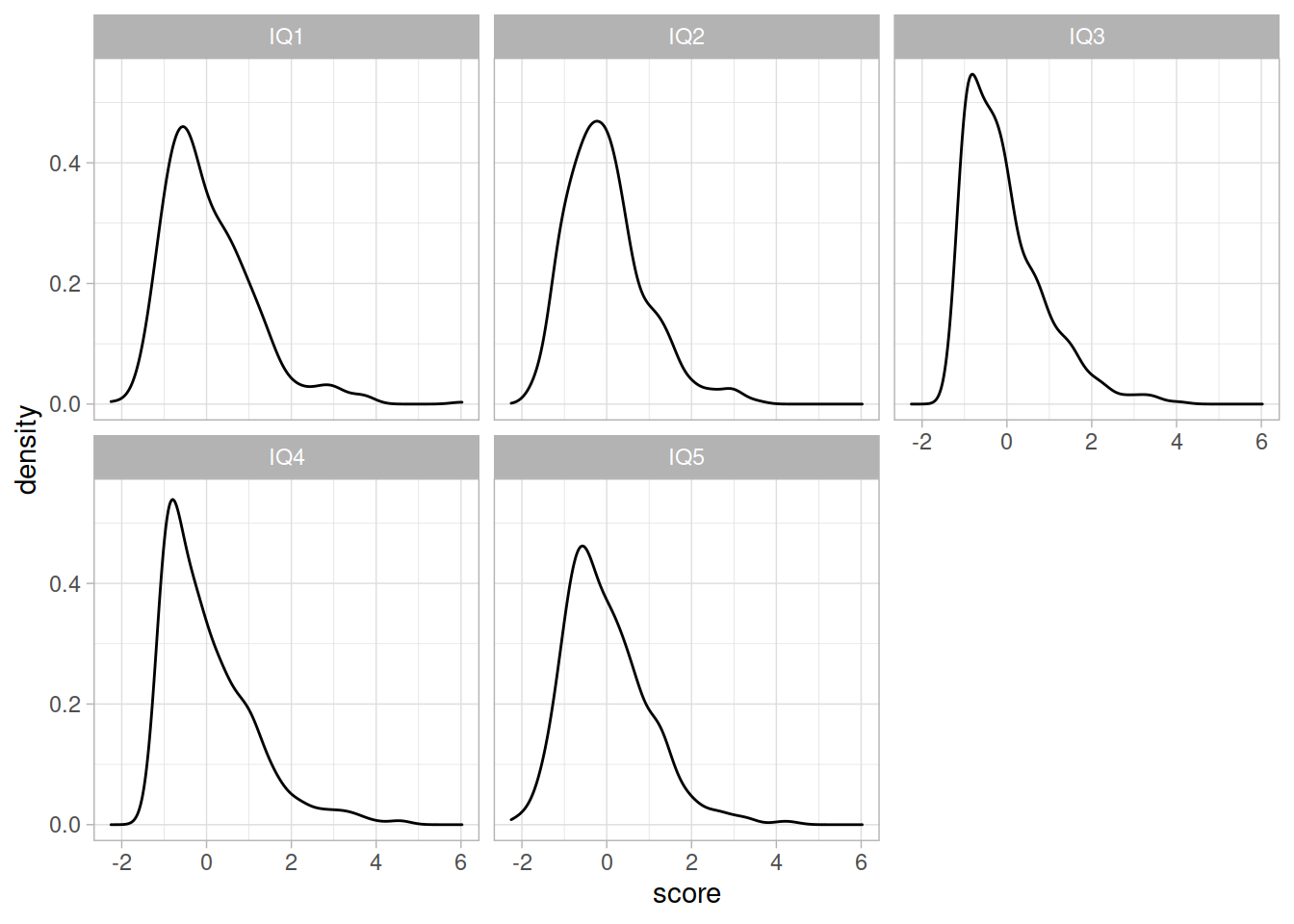
Now let’s conduct a CFA for the IQ items. Check their distributions (both numerically and visually) and fit a one-factor CFA using an appropriate estimation method.
Examine the fit of the CFA model. If necessary, check for areas of local misfit, and adjust your model accordingly.
Now its time to build a full SEM. Estimate the effect of prenatal stress on IQ.
Remember: We know that IQ is non-normal, so we would like to use a robust estimator (e.g. MLR). However, as lavaan will tell you if you try using estimator="MLR", this is not supported for ordered data (i.e., the Stress items). It suggests instead using the WLSMV (weighted least square mean and variance adjusted) estimator.
Specify and estimate your SEM model.
Missing data
Missingness
It is very common to have missing data. Participants may not stop halfway through the study, may decline to be followed up (if it is longitudinal) or may simply decline to answer certain sections. In addition, missing data can occur for all sorts of technical reasons (e.g, website crashed an auto-submits the questionnaire, etc.).
It is important to understand the possible reasons for missing data in order to appropriately consider what data you do have. If missing data are missing completely random, then the data you do have should still be representative of the population. But suppose you are studying cognitive decline in an aging population, and people who are suffering from cognitive impairment are less likely to attend the follow-up assessments. Is this missingness random? No. Does it affect how you should consider your available data to represent the population of interest? Yes.
There are three main explanations for missing data:
MCAR: Missing Completely At Random. Data which are MCAR are missing data for which the propensity for missingness is completely independent of any observed or unobserved variables. It is truly random if the data are missing or not.
MAR: Missing At Random. Data which are MAR are missing data for which the propensity for missingness is not random, but it can be fully explained by some variables for which there is complete data. In other words, there is a systematic relationship between missing values and observed values. For example, people who are unemployed at time of assessment will likely not respond to questions on job satisfaction. Missing values on job satisfaction is unrelated to the levels of job satisfaction, but related to their employment status.
MNAR: Missing Not At Random. Data which are MNAR are missing data for which the propensity for missingness is related to the value which is missing. For example, suppose an employer tells employees that there are a limited number of bonuses to hand out, and then employees are asked to fill out a questionnaire. Thos who are less satisfied with their job may not respond to questions on job satisfaction (if they believe it will negatively impact their chances of receiving a bonus).
To me, these are some of the most confusing terms in statistics, because “at random” is used to mean “not completely random!” It might be easier to think of “missing at random” as “missing conditionally at random,” but then it gives the same acronym as “completely at random!”
FIML (Full Information Maximum Likelihood)
One approach to dealing with missing data (not discussed in depth here) is imputation. This involves substituting missing values with values predicted by some model of the process which reflects the data generating process for that variable (sometimes including some stochasticity in these imputed values, sometimes not).1
In SEM, there is an extremely useful method known as full information maximum likelihood (FIML) which, similar way to imputation, uses observed data to supplement missingness. Intuitively, this is a bit like using all rest of the picture to fill in the missing bits (e.g. Figure 4).

Figure 4: Missingness
FIML utilises all observed variables (hence “full information”) to estimate missing values by maximising the likelihood of the sample as a function of the joint probability distribution of all variables. What is really neat about this is that it allows us to make full use of all our data, as it estimates missing values on all variables in our system, regardless of whether they are exogenous/endogenous variables. Compare this to functions such as lm() and lmer(), in which missing values on our explanatory variables result in simple list-wise deletion from the model. A downside, one could argue, is that you may have specific theoretical considerations about which observed variables should weigh in on estimating missingness in variable \(x\) (rather than all variables), in which case imputation techniques may be preferable.
In lavaan, we can make use of full information maximum likelihood by using missing = "FIML" in the functions cfa() and sem().
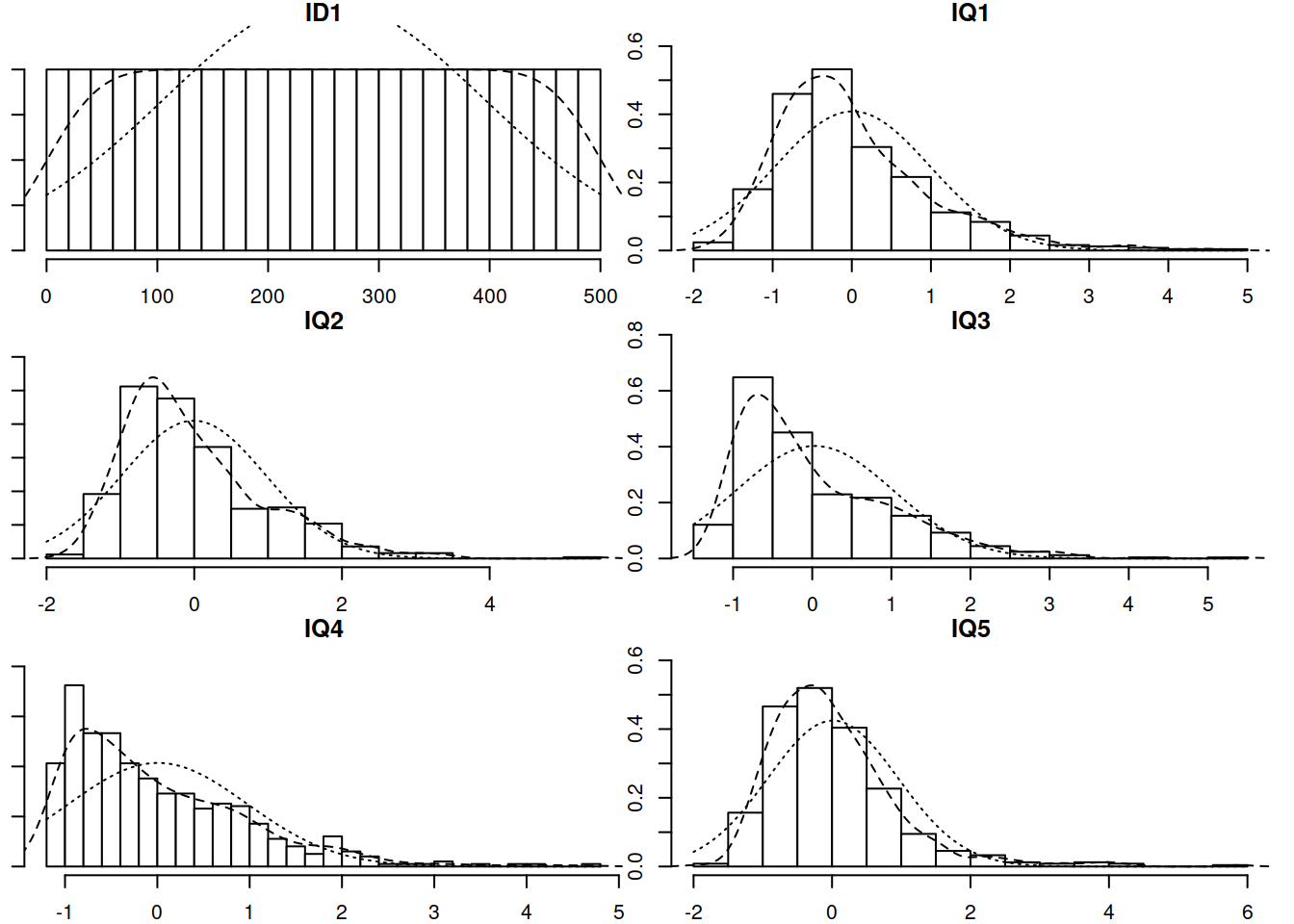
In order to try and replicate the IQ CFA, our researcher collects a new sample of size \(n=500\). However, she has some missing data. Specifically, those who scored poorly on earlier tests tended to feel discouraged and chose not to complete further tests.
Read in the new dataset, plot and numerically summarise the univariate distributions of the measured variables, and then conduct a CFA using the new data, taking account of the missingness (don’t forget to also use an appropriate estimator to account for any non-normality)
- The data can be found at https://uoepsy.github.io/data/IQdatam.csv, and is in .csv format.
Simulating SEM
simulating data as an aid to learning
An extremely useful approach to learning both R and statistics is to create yourself some fake data on which you can try things out. Because you create the data, you can control any relationships, group differences etc. In doing so, you can make yourself a target to aim for.
A note from Josiah & Umberto:
Many of you will currently be in the process of collecting/acquiring data for your thesis. If you are yet to obtain your data, we strongly recommend that you start to simulate some data with the expected distributions in order to play around and test how your analyses works, and how to interpret the results.
I would estimate that between us we generate fake data 5+ times a week on average, in order to help us work out things we don’t understand. It also enables for easy sharing of reproducible chunks of code which can perfectly replicate issues and help discussions.
We’re going to walk through the process of fitting a structural equation model by first simulating some data.
The data simulated was generated with the following parameters (expressed in lavaan syntax). You might describe this as the “data generating process.” The goal of statistics is to shed light on the data generating process when we don’t know what it is (when we have real data).
f1 =~ 0.8*item1 + 0.8*item2 + 0.5 * item3 + 0.5 * item4
f2 =~ 0.7*item5 + 0.8*item6 + 0.3 * item7 + 0.5 * item8
f3 =~ 0.4*item9 + 0.8*item10 + 0.4 * item11 + 0.5 * item12
f4 =~ 0.5*item13 + 0.5*item14 + 0.8*item15
f1 ~ .2*f2 + 0.8*f3 + f4
f2 ~ 0.7*f4
item3 ~~ 0.8*item4As you can see, in the data generating process, there are 4 factors each measured by 3 or 4 items. Factor f1 is regressed onto f2, f3 and f4, with f2 also regressed on to f4. We also specify a residual item covariance between items 3 and 4
You can find the data at https://uoepsy.github.io/data/simsem.csv, or run the code below to simulate it yourself (you’ll need to install the simstandard package).
set.seed(987)
mdl <- "
f1 =~ 0.8*item1 + 0.8*item2 + 0.5 * item3 + 0.5 * item4
f2 =~ 0.7*item5 + 0.8*item6 + 0.3 * item7 + 0.5 * item8
f3 =~ 0.4*item9 + 0.8*item10 + 0.4 * item11 + 0.5 * item12
f4 =~ 0.5*item13 + 0.5*item14 + 0.8*item15
f2 ~~ 0.3*f3
f1 ~ .2*f2 + 0.8*f3 + f4
f2 ~ 0.7*f4
item3 ~~ 0.8*item4
"
semPaths(lavaanify(mdl))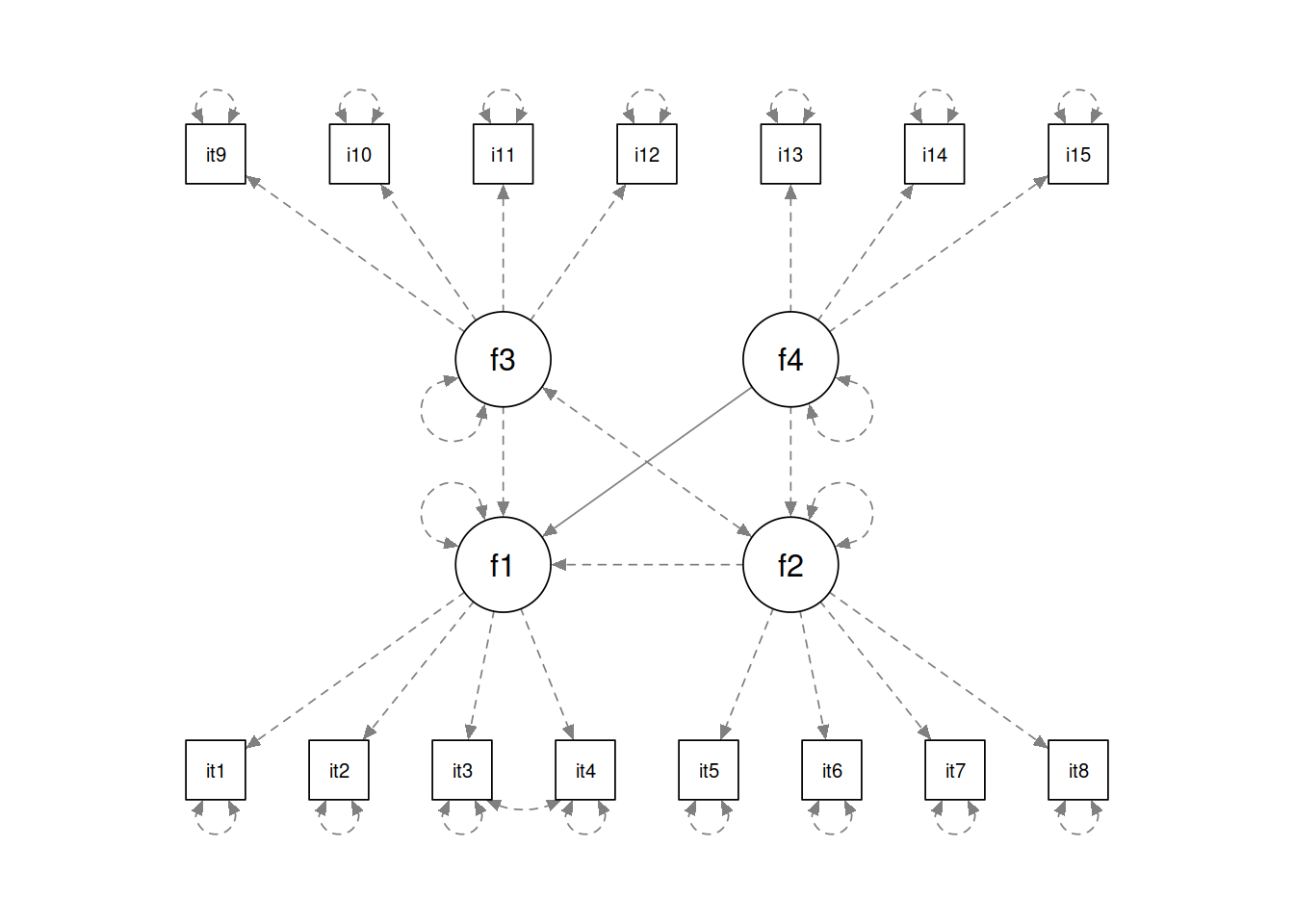
# from the simstandard package
df <- simstandard::sim_standardized(mdl, n = 1e3,
latent = FALSE,
errors = FALSE)Try playing around and fitting different models to the data, and evaluating how well they fit. What happens if you misspecify the measurement model? What happens if you leave out estimating the covariance between items3 and 4? Check your modification indices.
You will often see things like mean imputation and median imputation, but think carefully about why this might not be a great approach. It assumes that a single value is exactly the value which we would have observed if it were not missing. The lack of variability around this estimate of missing values will shrink the standard errors because it is assumed that no deviations exist among the substituted values.↩︎
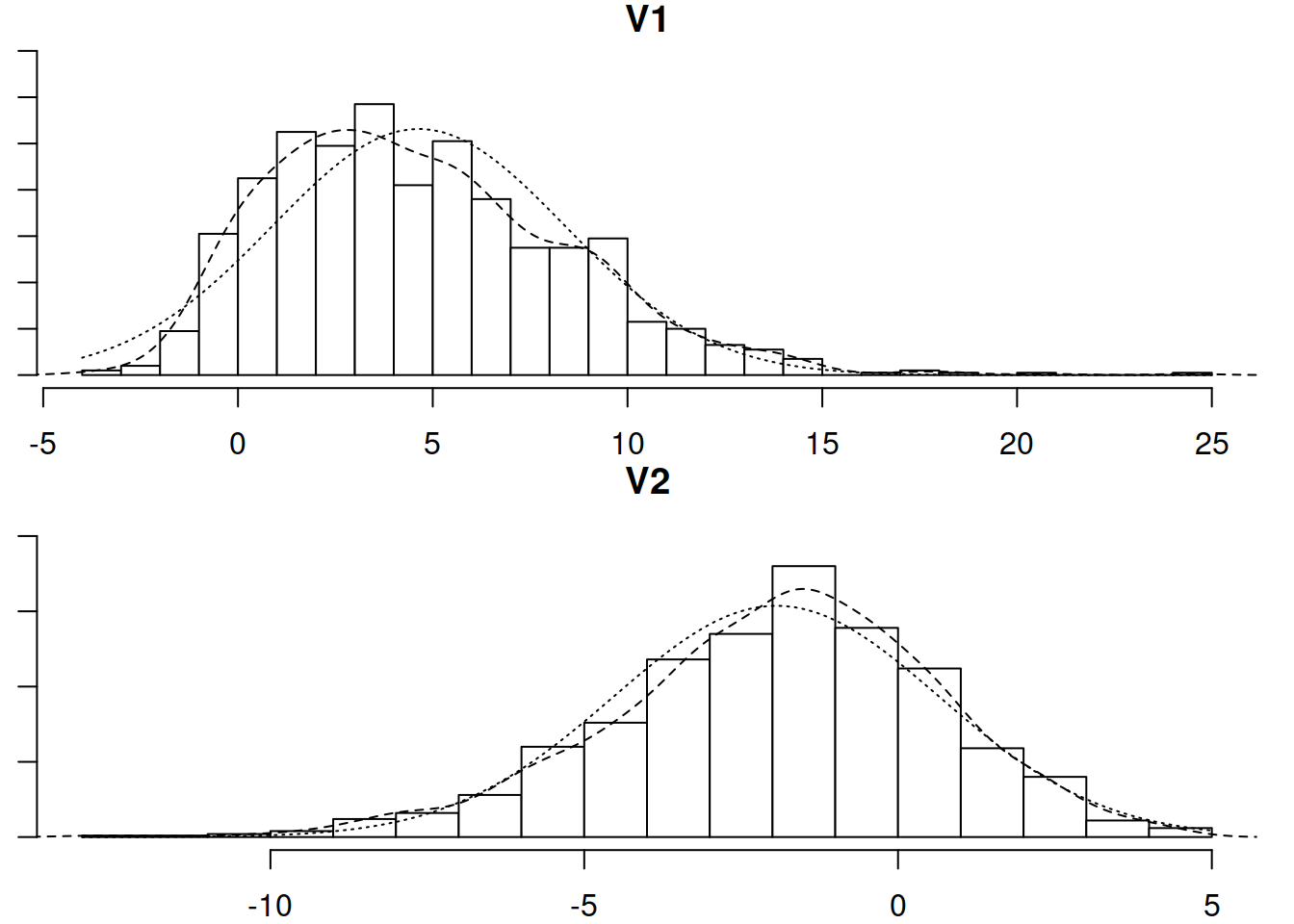 The joint probability distribution of these two variables can be visualised in Figure
The joint probability distribution of these two variables can be visualised in Figure