| QuestionNumber | Over the last 2 weeks, how much have you had/have you been... |
|---|---|
| item1 | Little interest or pleasure in doing things? |
| item2 | Feeling down, depressed, or hopeless? |
| item3 | Trouble falling or staying asleep, or sleeping too much? |
| item4 | Feeling tired or having little energy? |
| item5 | Poor appetite or overeating? |
| item6 | Feeling bad about yourself - or that you are a failure or have let yourself or your family down? |
| item7 | Trouble concentrating on things, such as reading the newspaper or watching television? |
| item8 | Moving or speaking so slowly that other people could have noticed? Or the opposite - being so fidgety or restless that you have been moving around a lot more than usual? |
| item9 | A lack of motivation to do anything at all? |
| item10 | Feeling nervous, anxious or on edge? |
| item11 | Not being able to stop or control worrying? |
| item12 | Worrying too much about different things? |
| item13 | Trouble relaxing? |
| item14 | Being so restless that it is hard to sit still? |
| item15 | Becoming easily annoyed or irritable? |
| item16 | Feeling afraid as if something awful might happen? |
W10 Exercises: EFA, replicability, reliability
More EFA
Data: petmoods2.csv
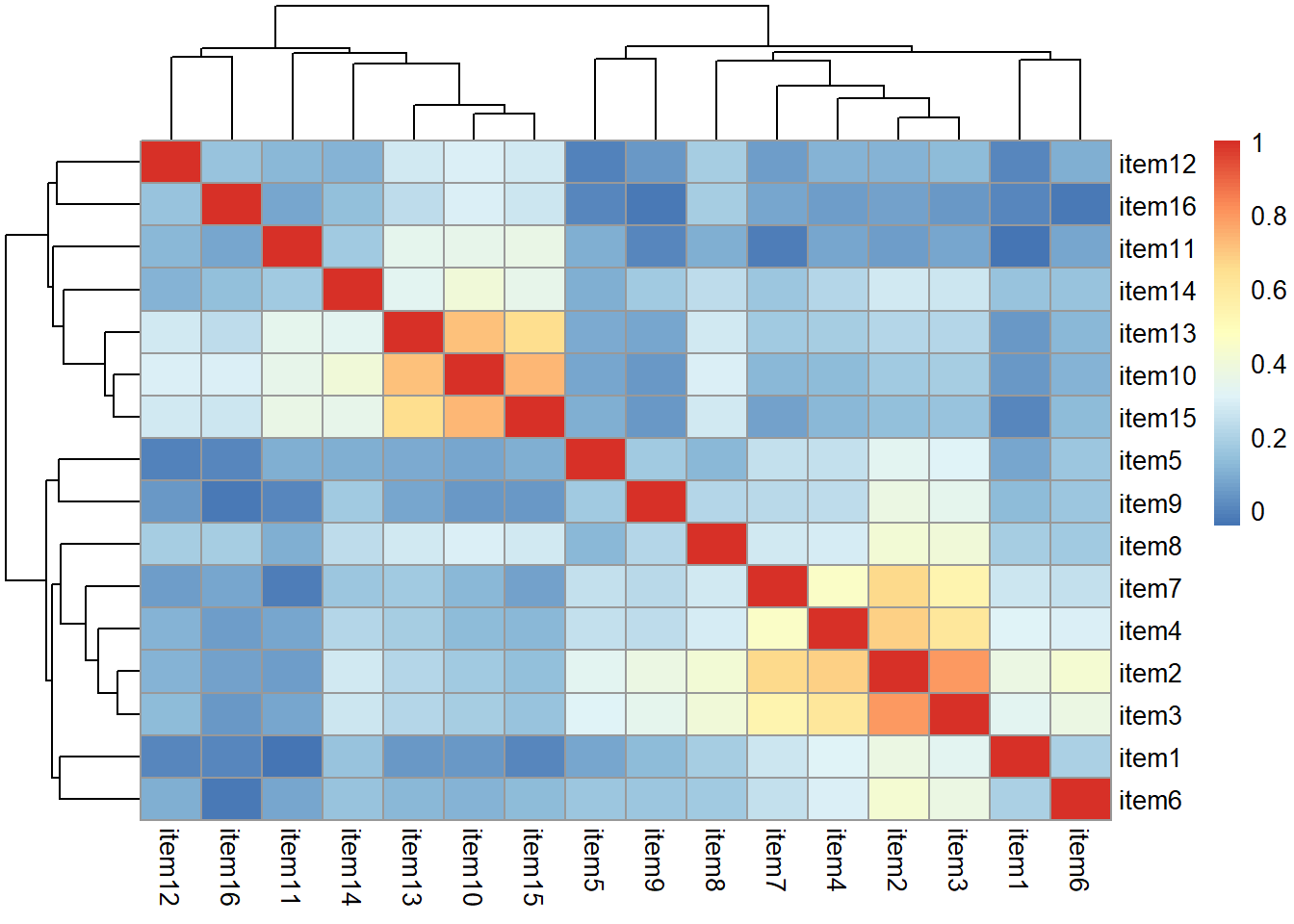
A pet food company has conducted a questionnaire on the internet (\(n = 620\)) to examine whether owning a pet influences low mood. They asked 16 questions on a Likert scale (1-7, detailed below) followed by a simple Yes/No question concerning whether the respondent owned a pet.
The researchers don’t really know much about the theory of mood disorders, but they looked at some other questionnaires and copied the questions they saw often appearing. They think that they are likely picking up on multiple different types of “low mood”, so they want do an EFA to examine this. They then want to look at group differences (pet owners vs not pet owners) in the dimensions of low mood.
The data are available at https://uoepsy.github.io/data/petmoods2.csv
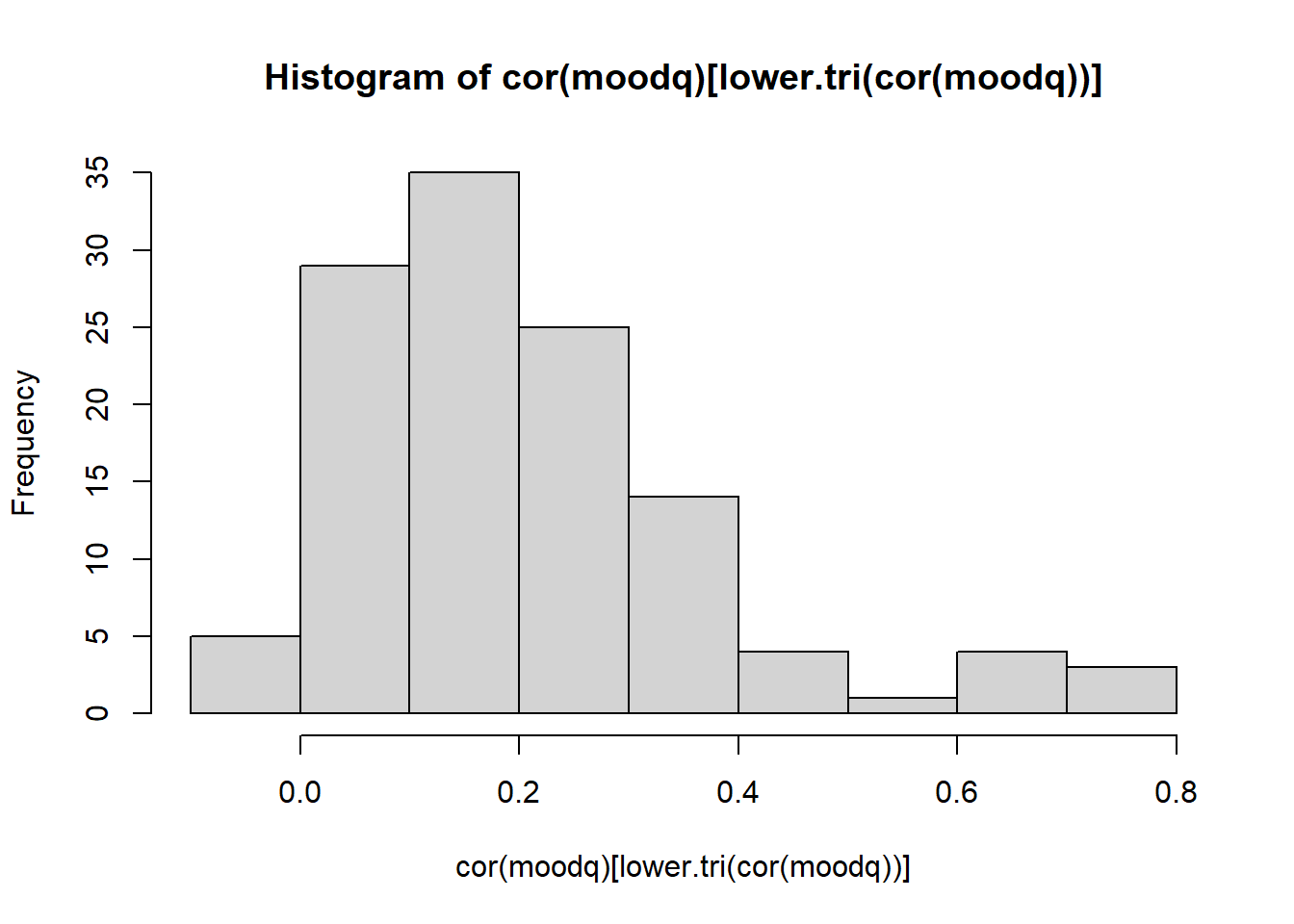
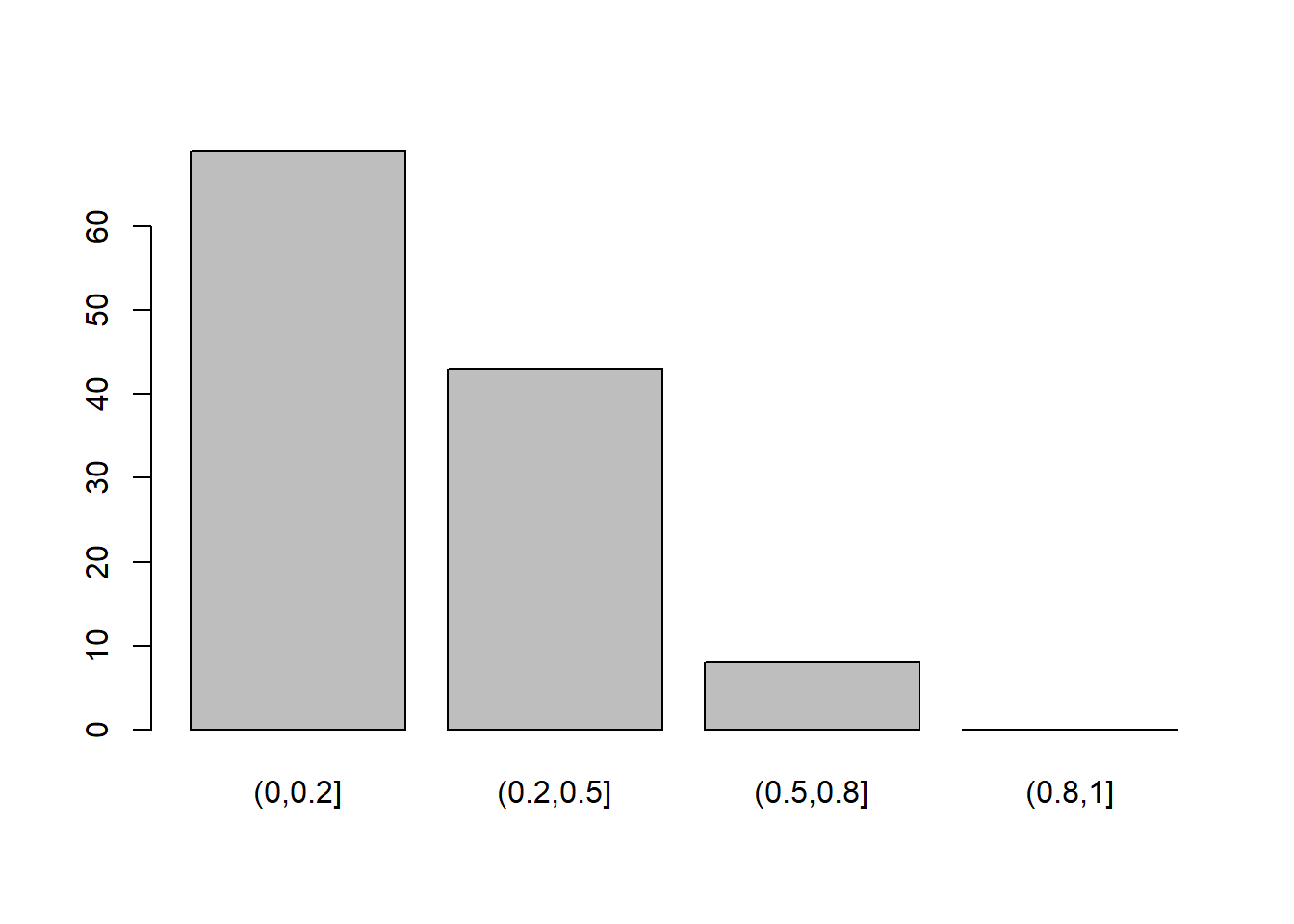
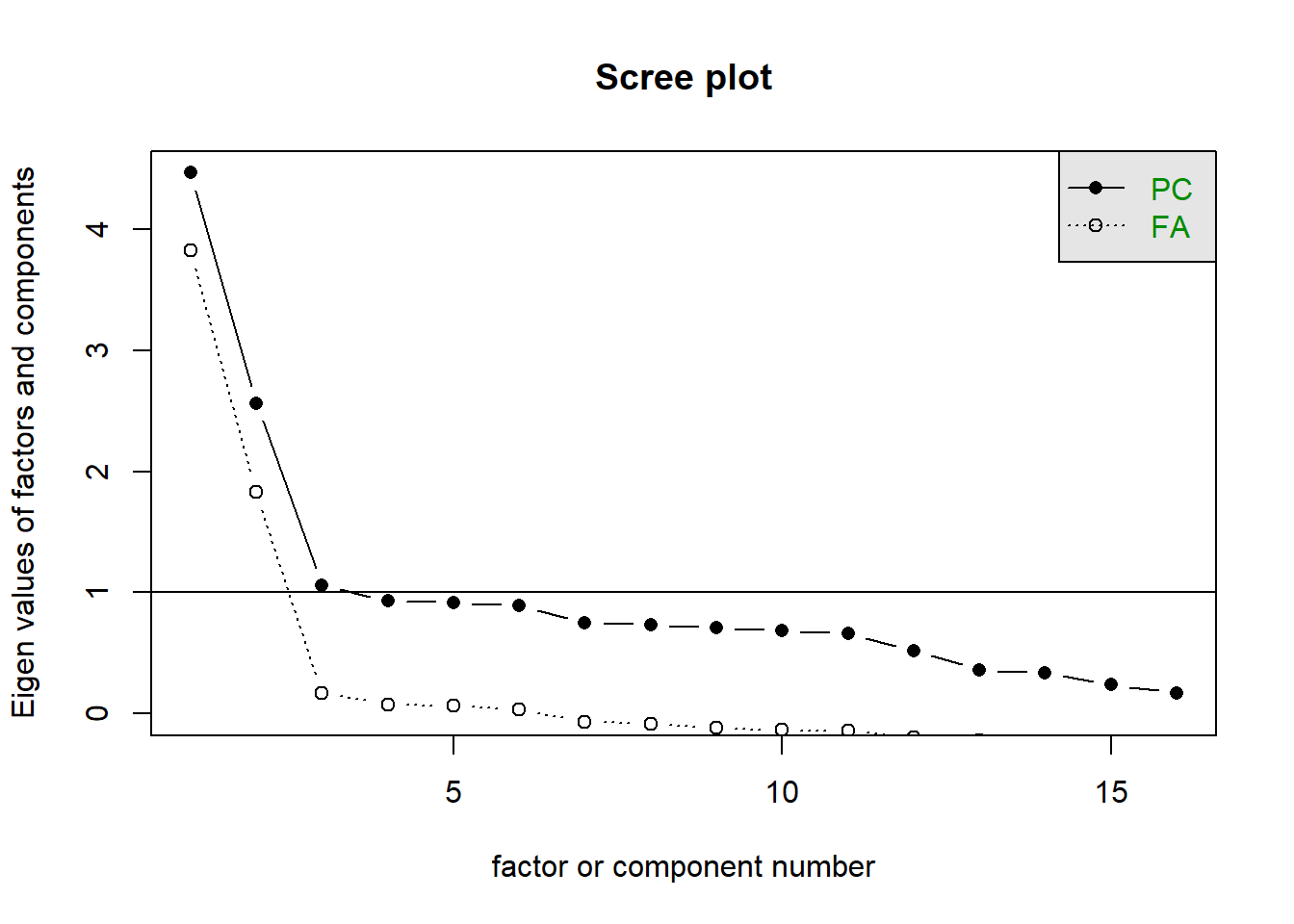
Read in the data, check the suitability for conducting factor analysis, and then examine what underlying dimensions best explain the observed relationships between the 16 mood-related questions in the survey.
- It will be very handy to rename the columns to something easier to work with (see R7-Questionnaire Data Wrangling #variable-names).
- Make sure to fit the EFA on only the mood questions
Replicability
Splitting the dataset in two, calculate congruence coefficients for your factor solution.
The most reliable way to split a dataset is actually to create a sample of rows.
For example, if I had 100 rows, then I can split it into two groups of 50 using:
idx <- sample(1:100, 50) # sample 50 rows
split1 <- data[idx, ] # choose those rows
split2 <- data[-idx, ] # exclude those rowsTo calculate congruence coefficients, fit the same factor solution to both datasets and then use fa.congruence()
Ideally, we would split a dataset in two right at the start, develop our model on the “exploratory” half, and not touch the second half of the data until we want to assess congruence.
If we had unlimited time and resources, we would just collect another, completely independent, sample.
So let’s pretend that’s exactly what we’ve done!
You can find a 2nd dataset at https://uoepsy.github.io/data/petmoods_conf.csv that contains the same questions.
Compute congruence coefficients for your factor solution across the two dataset (the first one with n=620 and this one with n=203).
Reliability
Calculate two measures of reliability for each factor - \(alpha\) and \(omega\). How do they differ?
(If you’re thinking “which one should I use?” then there’s not really a right answer - they rely on assuming different measurement models. If you’re going to use mean/sum scores, then reporting reliabilty as \(\alpha\) will make more sense)
Make sure to do this separately for each factor, because both \(\alpha\) and \(\omega_{total}\) assume unidimensionality
Getting Scores
We’re going to demonstrate how these decisions can have a bearing on your analysis.
Remember, we’re interested in ultimately doing a group comparison (pet owners vs non-pet owners) in the dimension(s) of low mood.
So we’re going to need numbers to give each person’s standing on ‘low mood’ dimension(s).
For each of your factors, create two scores: first calculate a sum score from the relevant items, and then estimate a factor score using factor.scores
- We’re essentially asking
low mood ~ petownership, so the dimensions of low mood are dependent variables. So use the Bartlett method of estimating scores.
- You’ll probably want to append them to the end of the original dataset where we have the variable about pet ownership.
Conduct a \(t\)-test to examine whether the pet-owners differ from non-pet-owners in their levels of each factor of low mood.
Try doing this with the sum scores, and then with the factor scores. What is different?
(If you’re wondering “which one is right?” then the answer is kind of “both”/“neither”! Just like \(\alpha\) and \(\omega\), they are assuming different measurement models of the two dimensions)