Assumptions & Diagnostics: The Recipe Book
Helpful Mnemonics
It may help to think of the sequence of steps involved in statistical modeling as:
\[
\text{Choose} \rightarrow \text{Fit} \rightarrow \text{Assess} \rightarrow \text{Use}
\]
- We explore/visualise our data and Choose our model specification.
- Then we Fit the model in R.
- Next, we Assess the fit, to ensure that it meets all the underlying assumptions?
- Finally, we Use our model to draw statistical inferences about the world, or to make predictions.
The assumptions of the linear model can be committed to memory using the LINE mnemonic:
- Linearity: The relationship between \(y\) and \(x\) is linear.
- Independence of errors: The error terms should be independent from one another.
- Normality: The errors \(\epsilon\) are normally distributed
- Equal variances (“Homoscedasticity”): The scale of the variability of the errors \(\epsilon\) is constant at all values of \(x\).
When we fit a model, we evaluate many of these assumptions by looking at the residuals
(the deviations from the observed values \(y_i\) and the model estimated value \(\hat y_i\)).
The residuals, \(\hat \epsilon\) are our estimates of the actual unknown true error term \(\epsilon\). These assumptions hold both for a regression model with a single predictor and for one with multiple predictors.
Setup
:::
For this guide, we are going to use the following model:
\[ \text{Wellbeing} = b_0 + b_1 \cdot \text{Outdoor Time} + b_2 \cdot \text{Social Interactions} + \epsilon \]
Which we fitted in R as follows:
library(tidyverse)
# Read in data
mwdata = read_csv(file = "https://uoepsy.github.io/data/wellbeing.csv")
# fit the model
wbmodel <- lm(wellbeing ~ outdoor_time + social_int, data = mwdata):::
Checking Each Assumption
Linearity
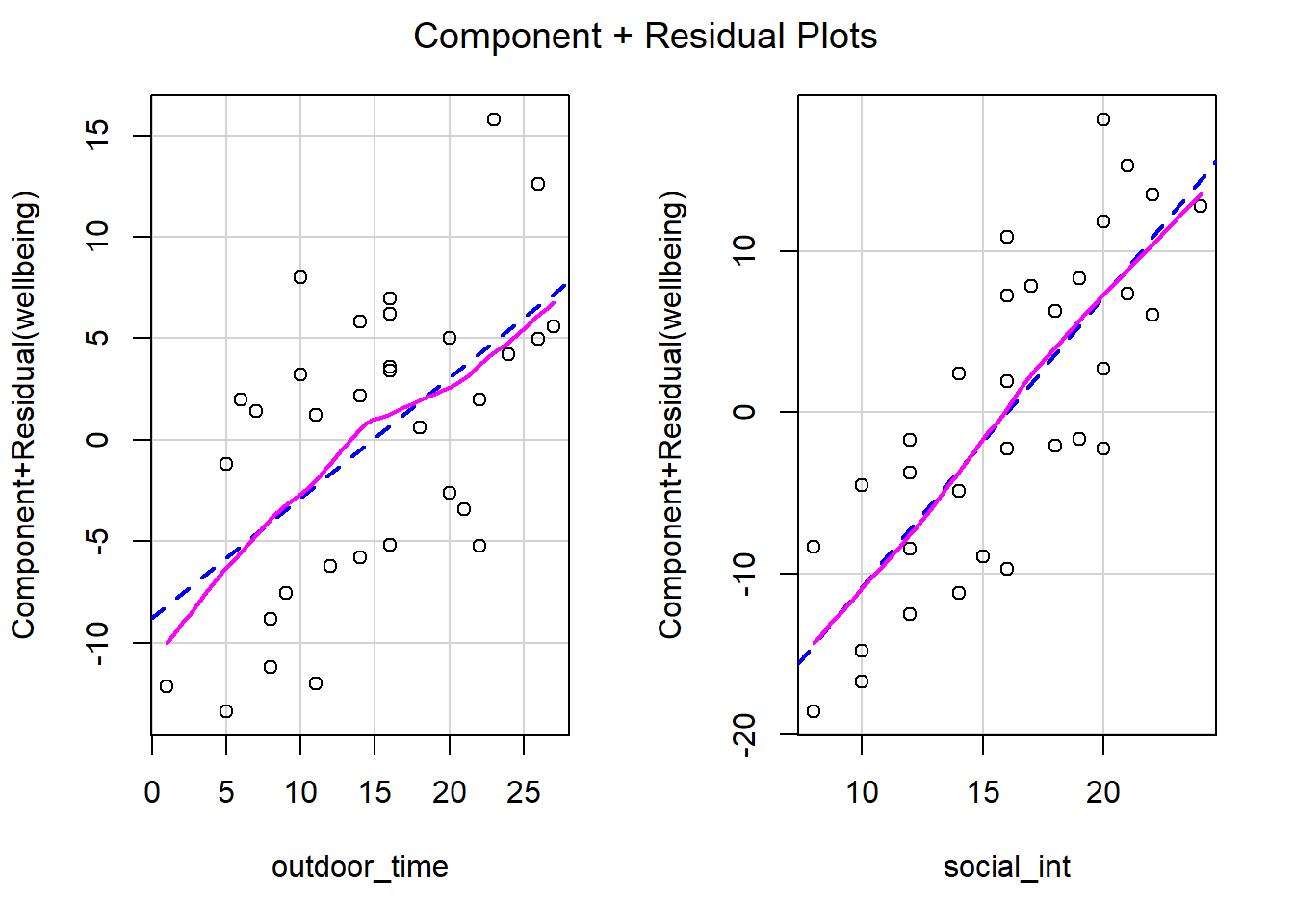
In simple linear regression with only one explanatory variable, we can assess linearity through a simple scatterplot of the outcome variable against the explanatory. In multiple regression, however, it becomes more necessary to rely on diagnostic plots of the model residuals. This is because we need to know whether the relations are linear between the outcome and each predictor after accounting for the other predictors in the model.
In order to assess this, we use partial-residual plots (also known as ‘component-residual plots’). This is a plot with each explanatory variable \(x_j\) on the x-axis, and partial residuals on the y-axis.
Partial residuals for a predictor \(x_j\) are calculated as: \[ \hat \epsilon + \hat b_j x_j \]
In R we can easily create these plots for all predictors in the model by using the crPlots() function from the car package.
Create partial-residual plots for the wbmodel model.
Remember to load the car package first. If it does not load correctly, it might mean that you have need to install it.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to the plots.
Equal variances (Homoscedasticity)
The equal variances assumption is that the error variance \(\sigma^2\) is constant across values of the predictors \(x_1\), … \(x_k\), and across values of the fitted values \(\hat y\). This sometimes gets termed “Constant” vs “Non-constant” variance. Figures 1 & 2 shows what these look like visually.
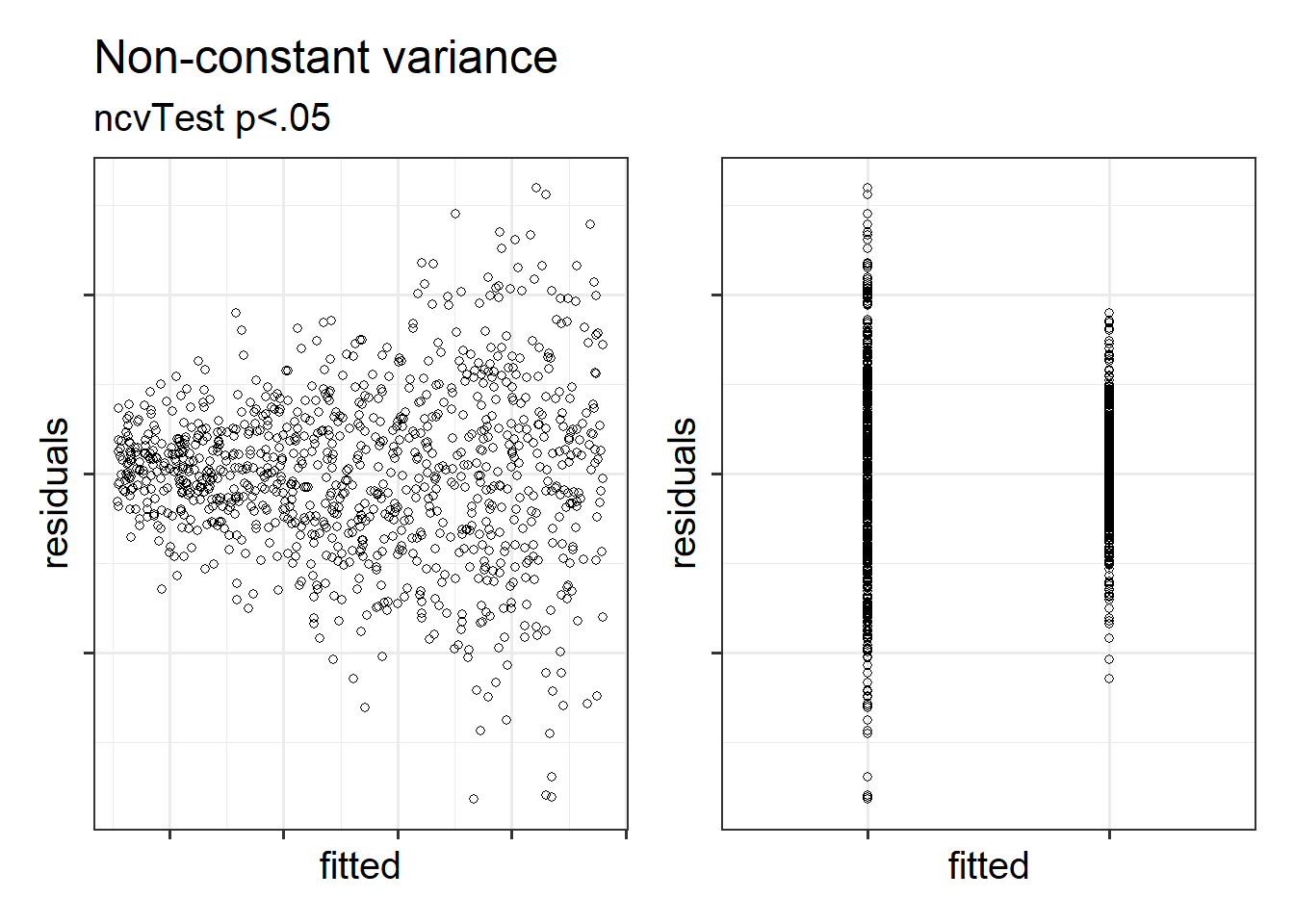
Figure 1: Non-constant variance for numeric and categorical x
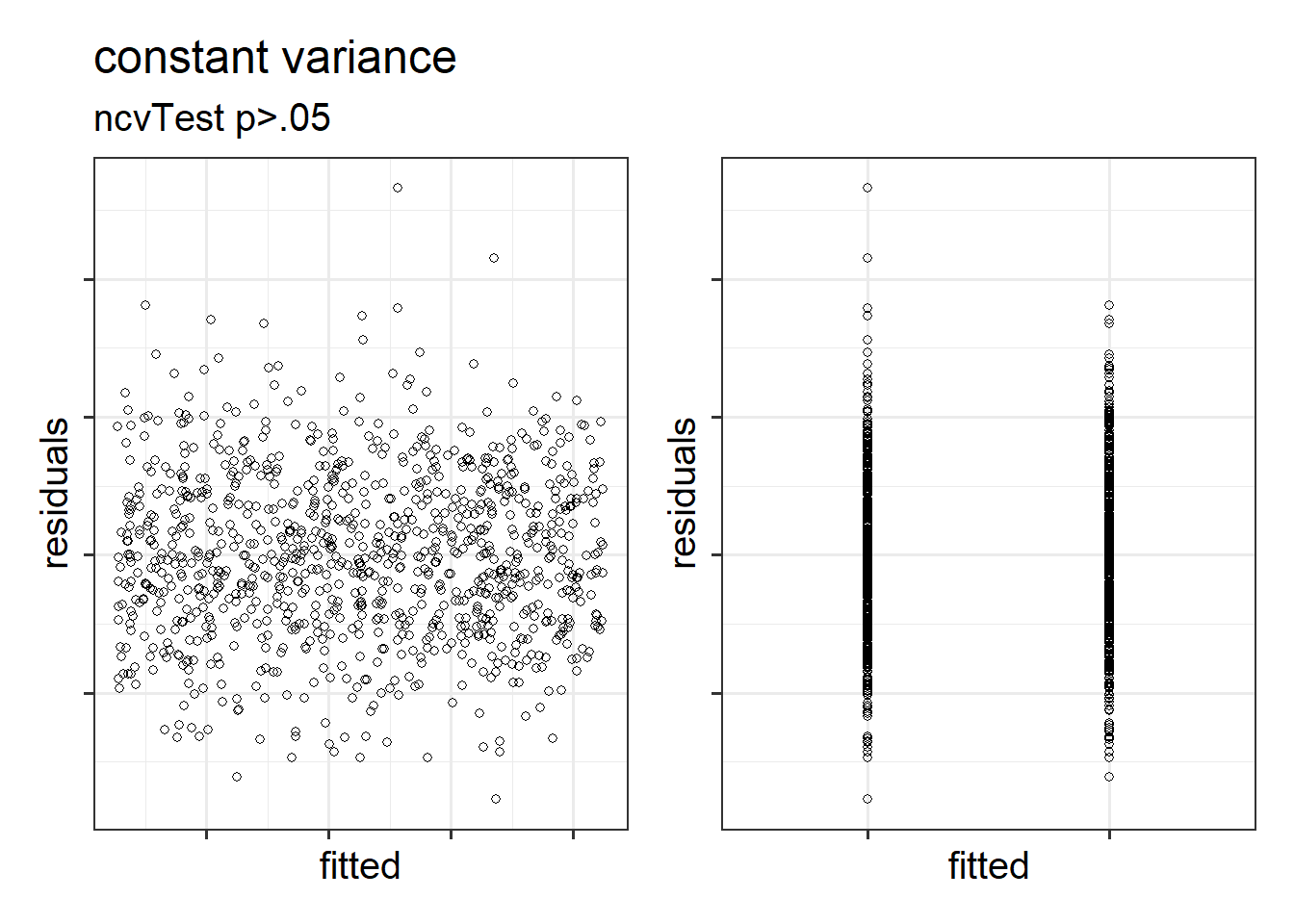
Figure 2: Constant variance for numeric and categorical x
In R we can create plots of the Pearson residuals against the predicted values \(\hat y\) and against the predictors \(x_1\), … \(x_k\) by using the residualPlots() function from the car package. This function also provides the results of a lack-of-fit test for each of these relationships (note when it is the fitted values \(\hat y\) it gets called “Tukey’s test”).
ncvTest(model) (also from the car package) performs a test against the alternative hypothesis that the error variance changes with the level of the fitted value (also known as the “Breusch-Pagan test”). \(p >.05\) indicates that we do not have evidence that the assumption has been violated.
Use residualPlots() to plot residuals against each predictor, and use ncvTest() to perform a test against the alternative hypothesis that the error variance changes with the level of the fitted value.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to plots and/or formal tests where available.
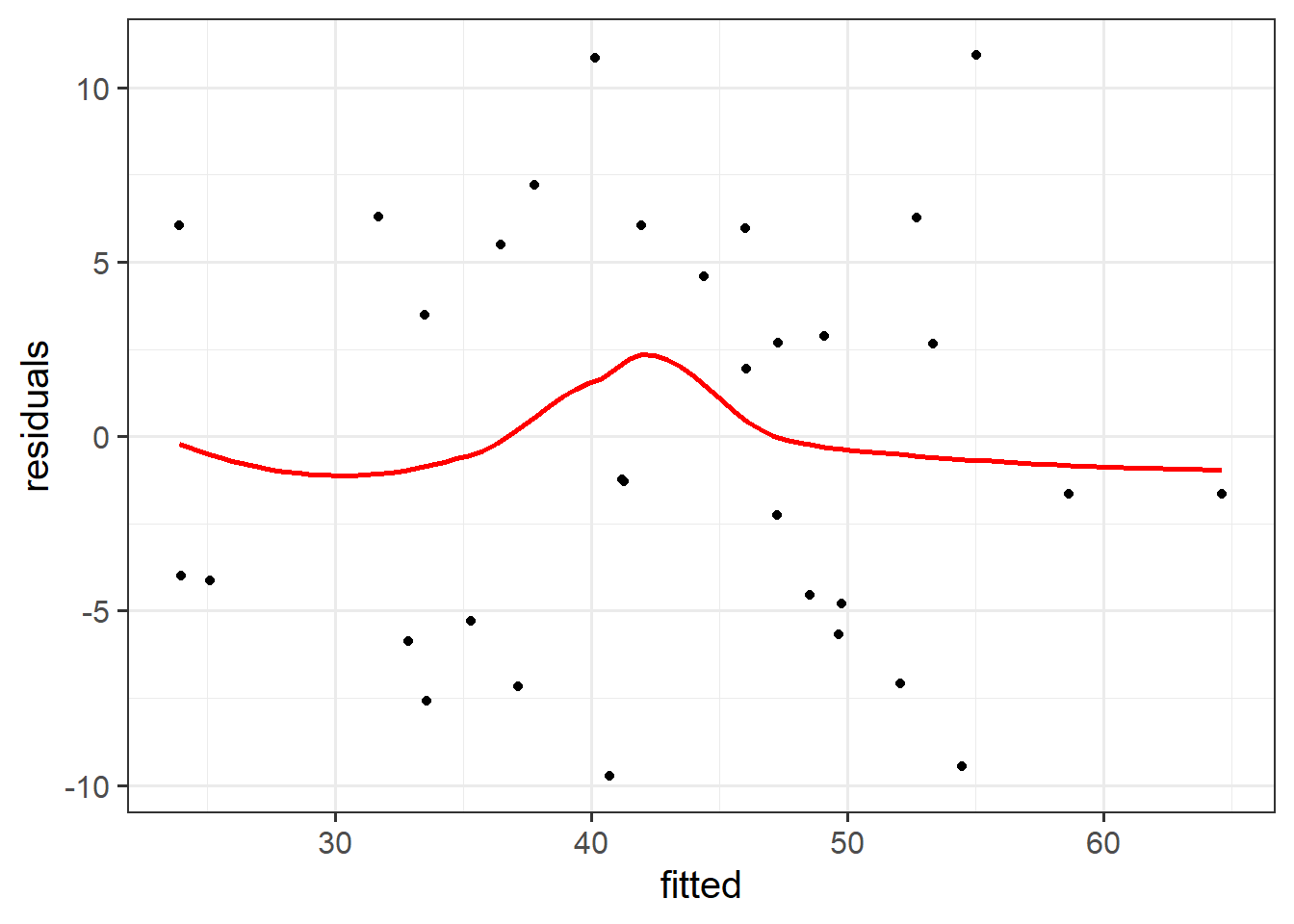
Create the “residuals vs. fitted plot” - a scatterplot with the residuals \(\hat \epsilon\) on the y-axis and the fitted values \(\hat y\) on the x-axis.
You can either do this:
- manually, using the functions
residuals()andfitted(), or - quickly by giving the
plot()function your model. This will actually give you lots of plots, so we can specify which plot we want to return - e.g.,plot(wbmodel, which = 1)
You can use this plot to visually assess:
- Linearity: Does the average of the residuals \(\hat \epsilon\) remain close to 0 across the plot?
- Equal Variance: does the spread of the residuals \(\hat \epsilon\) remain constant across the predicted values \(\hat y\)?
Independence
The “independence of errors” assumption is the condition that the errors do not have some underlying relationship which is causing them to influence one another.
There are many sources of possible dependence, and often these are issues of study design. For example, we may have groups of observations in our data which we would expect to be related (e.g., multiple trials from the same participant). Our modelling strategy would need to take this into account.
One form of dependence is autocorrelation - this is when observations influence those adjacent to them. It is common in data for which time is a variable of interest (e.g, the humidity today is dependent upon the rainfall yesterday).
In R we can test against the alternative hypothesis that there is autocorrelation in our errors using the durbinWatsonTest() (an abbreviated function dwt() is also available) in the car package.
Perform a test against the alternative hypothesis that there is autocorrelation in the error terms.
Write a sentence summarising whether or not you consider the assumption of independence to have been met (you may have to assume certain aspects of the study design).
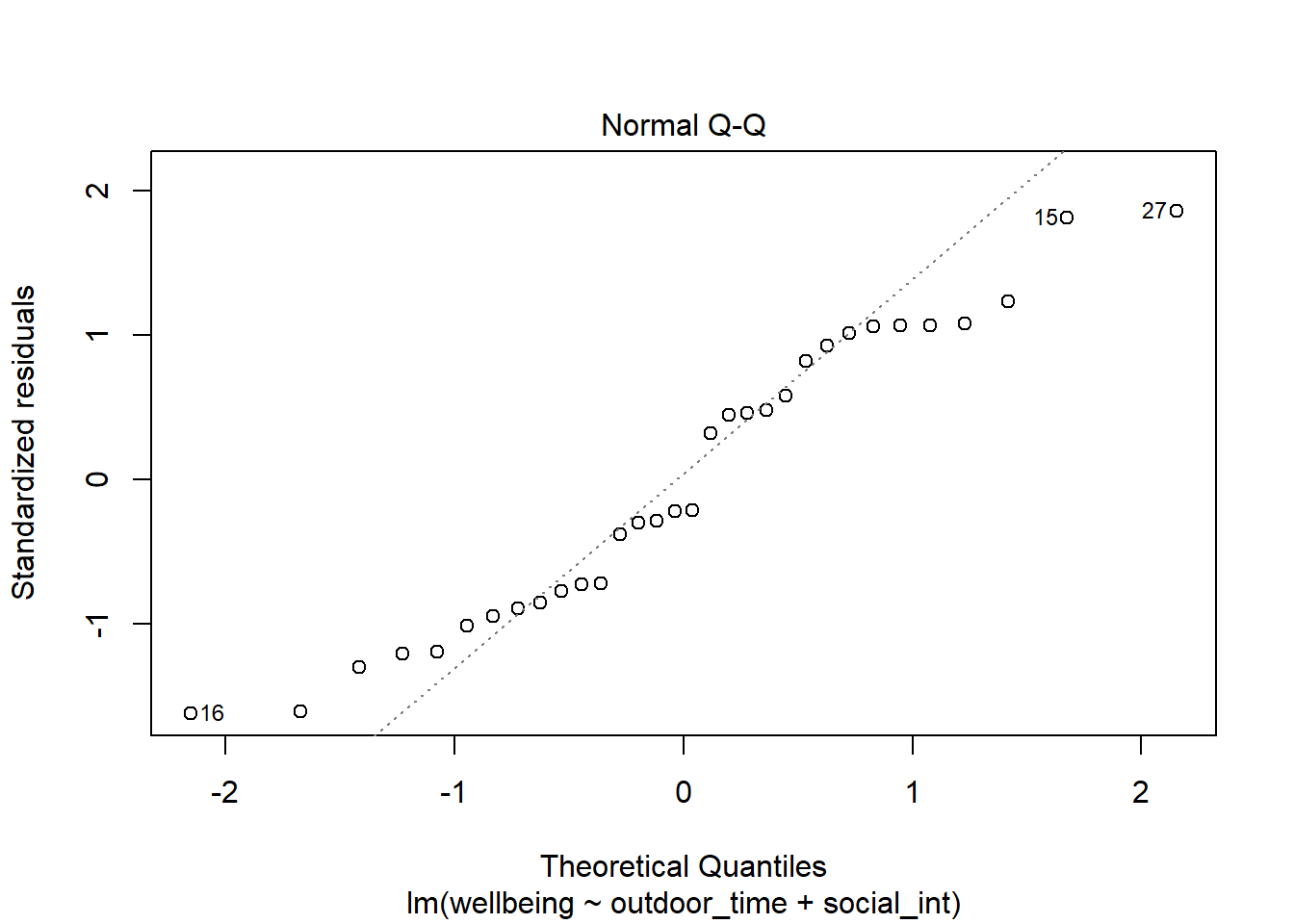
Normality of errors
The normality assumption is the condition that the errors \(\epsilon\) are normally distributed.
We can visually assess this condition through histograms, density plots, and quantile-quantile plots (QQplots) of our residuals \(\hat \epsilon\).
We can also perform a Shapiro-Wilk test against the alternative hypothesis that the residuals were not sampled from a normally distributed population.
- The
shapiro.test()function in R performs a Shapiro-Wilk test. plot(model_name, which = 2)gives us a QQplot of the residuals (or you can do it manually by extracting the residuals usingresid(model_name)).
Assess the normality assumption by producing a qqplot of the residuals (either manually or using plot(model, which = ???)), and conducting a Shapiro-Wilk test.
Write a sentence summarising whether or not you consider the assumption to have been met. Justify your answer with reference to plots and/or formal tests where available.
Multicollinearity
Recall our interpretation of multiple regression coefficients as
This interpretation falls down if predictors are highly correlated because if, e.g., predictors \(x_1\) and \(x_2\) are highly correlated, then changing the value of \(x_1\) necessarily entails a change the value of \(x_2\) meaning that it no longer makes sense to talk about holding \(x_2\) constant.
We can assess multicollinearity using the variance inflation factor (VIF), which for a given predictor \(x_j\) is calculated as:
\[
VIF_j = \frac{1}{1-R_j^2} \\
\]
Where \(R_j^2\) is the coefficient of determination (the R-squared) resulting from a regression of \(x_j\) on to all the other predictors in the model (\(x_j = x_1 + ... x_k + \epsilon\)).
The more highly correlated \(x_j\) is with other predictors, the bigger \(R_j^2\) becomes, and thus the bigger \(VIF_j\) becomes.
The square root of VIF indicates how much the SE of the coefficient has been inflated due to multicollinearity. For example, if the VIF of a predictor variable were 4.6 (\(\sqrt{4.6} = 2.1\)), then the standard error of the coefficient of that predictor is 2.1 times larger than if the predictor had zero correlation with the other predictor variables. Suggested cut-offs for VIF are varied. Some suggest 10, others 5. Define what you will consider an acceptable value prior to calculating it.
In R, the vif() function from the car package will provide VIF values for each predictor in your model.
Calculate the variance inflation factor (VIF) for the predictors in the model.
Write a sentence summarising whether or not you consider multicollinearity to be a problem here.
Individual Case Diagnostics
In linear regression, individual cases in our data can influence our model more than others. There are a variety of measures we can use to evaluate the amount of misfit and influence that single observations have on our model and our model estimates.
THERE ARE NO HARD RULES FOR WHAT COUNTS AS “INFLUENTIAL” AND HOW WE SHOULD DEAL WITH THESE CASES
There are many ways to make a cake. recipes can be useful, but you really need to think about what ingredients you actually have (what data you have).
You don’t have to exclude influential observations. Try to avoid blindly following cut-offs, and try to think carefully about outliers and influential points and whether you want to exclude them, and whether there might be some other model specification that captures this in some estimable way. Do these observations change the conclusions you make (you can try running models with and without certain cases).
There are various measures of outlyngness and influence. Here are a few:
- Regression outliers: A large residual \(\hat \epsilon_i\) - i.e., a big discrepancy between their predicted y-value and their observed y-value.
- Standardised residuals: For residual \(\hat \epsilon_i\), divide by the estimate of the standard deviation of the residuals. In R, the
rstandard()function will give you these - Studentised residuals: For residual \(\hat \epsilon_i\), divide by the estimate of the standard deviation of the residuals excluding case \(i\). In R, the
rstudent()function will give you these. Values \(>|2|\) (greater in magnitude than two) are considered potential outliers.
- Standardised residuals: For residual \(\hat \epsilon_i\), divide by the estimate of the standard deviation of the residuals. In R, the
- High leverage cases: These are cases which have considerable potential to influence the regression model (e.g., cases with an unusual combination of predictor values).
- Hat values: are used to assess leverage. In R, The
hatvalues()function will retrieve these.
Hat values of more than \(2 \bar{h}\) (2 times the average hat value) are considered high leverage. \(\bar{h}\) is calculated as \(\frac{k + 1}{n}\), where \(k\) is the number of predictors, and \(n\) is the sample size.
- Hat values: are used to assess leverage. In R, The
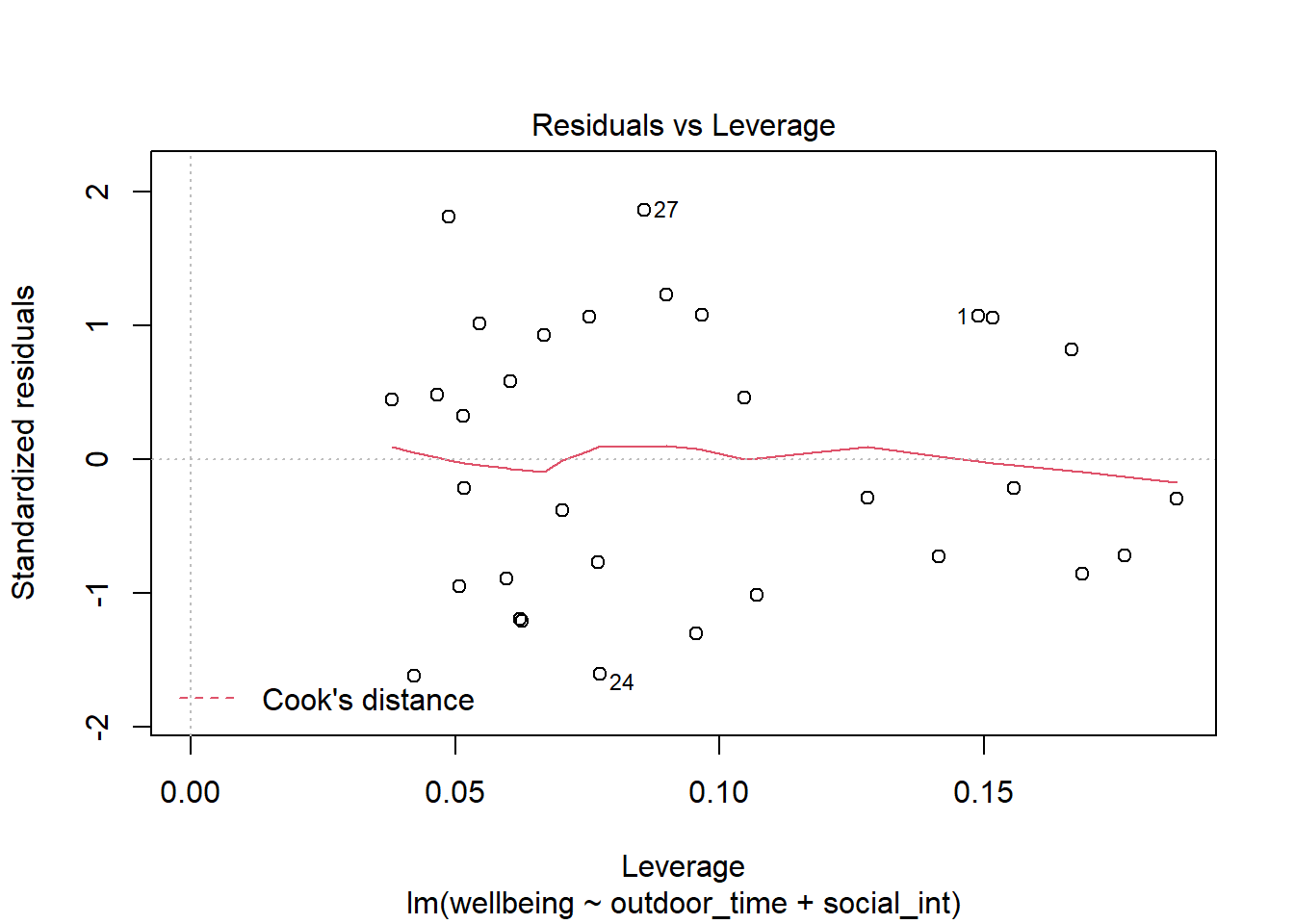
- High influence cases: When a case has high leverage and is an outlier, it will have a large influence on the regression model.
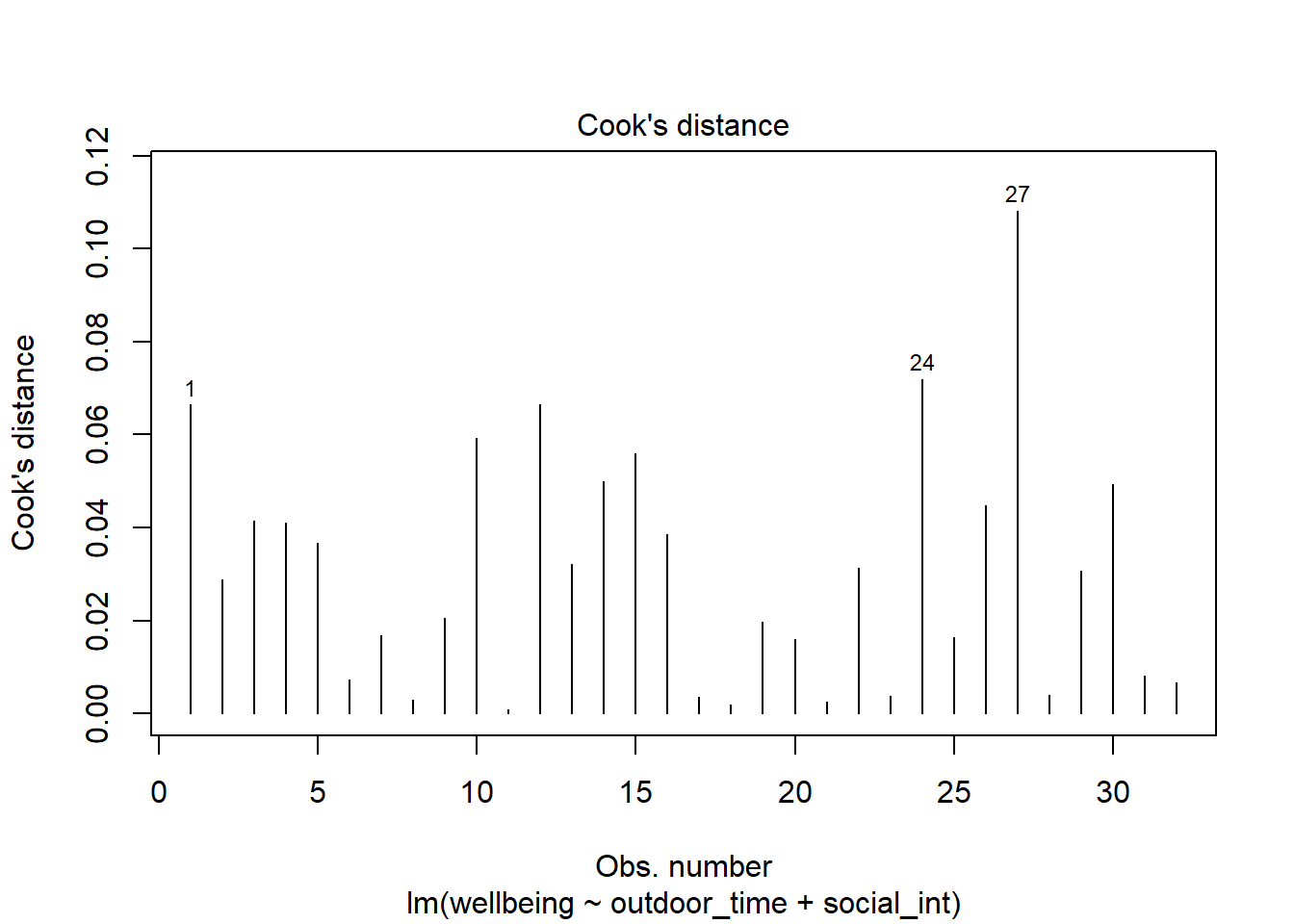
- Cook’s Distance: combines leverage (hatvalues) with outlying-ness to capture influence. In R, the
cooks.distance()function will provide these.
There are many suggested Cook’s Distance cut-offs.
- Cook’s Distance: combines leverage (hatvalues) with outlying-ness to capture influence. In R, the
Create a new tibble which contains:
- The original variables from the model (Hint, what does
<fitted model>$modelgive you?) - The fitted values from the model \(\hat y\)
- The residuals \(\hat epsilon\)
- The studentised residuals
- The hat values
- The Cook’s Distance values.
Looking at the studentised residuals, are there any extreme values?
Looking at the hat values, are there any observations with high leverage?
Plot the Cook’s Distance values, does it look like there may be any highly influential points?
(You can use plot(model, which = 4) and plot(model, which = 5)).
Other influence.measures()
Alongside Cook’s Distance, we can examine the extent to which model estimates and predictions are affected when an entire case is dropped from the dataset and the model is refitted.
- DFFit: the change in the predicted value at the \(i^{th}\) observation with and without the \(i^{th}\) observation is included in the regression.
- DFbeta: the change in a specific coefficient with and without the \(i^{th}\) observation is included in the regression.
- DFbetas: the change in a specific coefficient divided by the standard error, with and without the \(i^{th}\) observation is included in the regression.
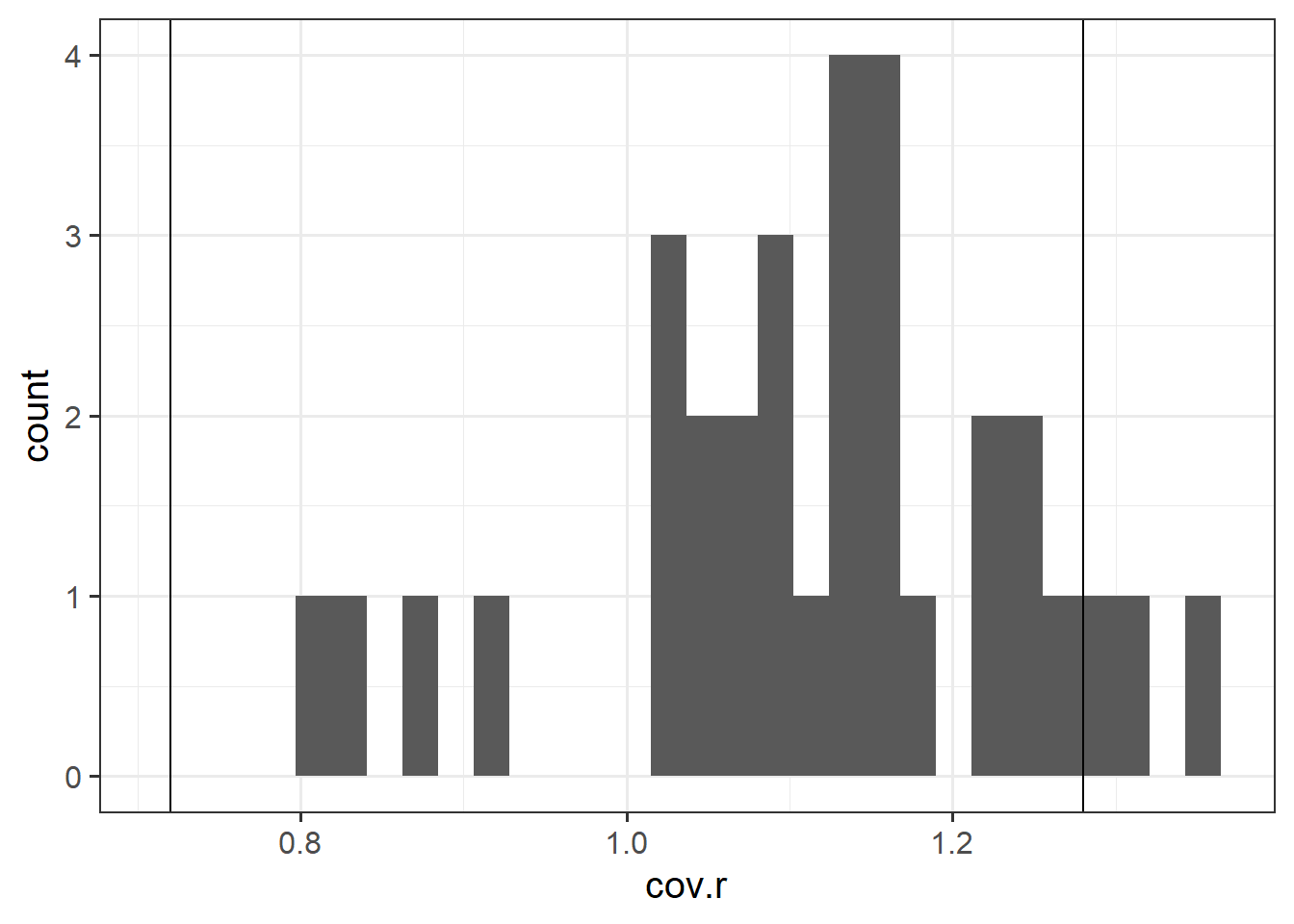
- COVRATIO: measures the effect of an observation on the covariance matrix of the parameter estimates. In simpler terms, it captures an observation’s influence on standard errors. Values which are \(>1+\frac{3(k+1)}{n}\) or \(<1-\frac{3(k+1)}{n}\) are sometimes considered as having strong influence.
Use the function influence.measures() to extract these delete-1 measures of influence.
Try plotting the distributions of some of these measures.
Tip: the function influence.measures() returns an infl-type object. To plot this, we need to find a way to extract the actual numbers from it.
What do you think names(influence.measures(<fitted model>)) shows you? How can we use influence.measures(<fitted model>)$ ???? to extract the matrix of numbers?

This workbook was written by Josiah King, Umberto Noe, and Martin Corley, and is licensed under a Creative Commons Attribution 4.0 International License.
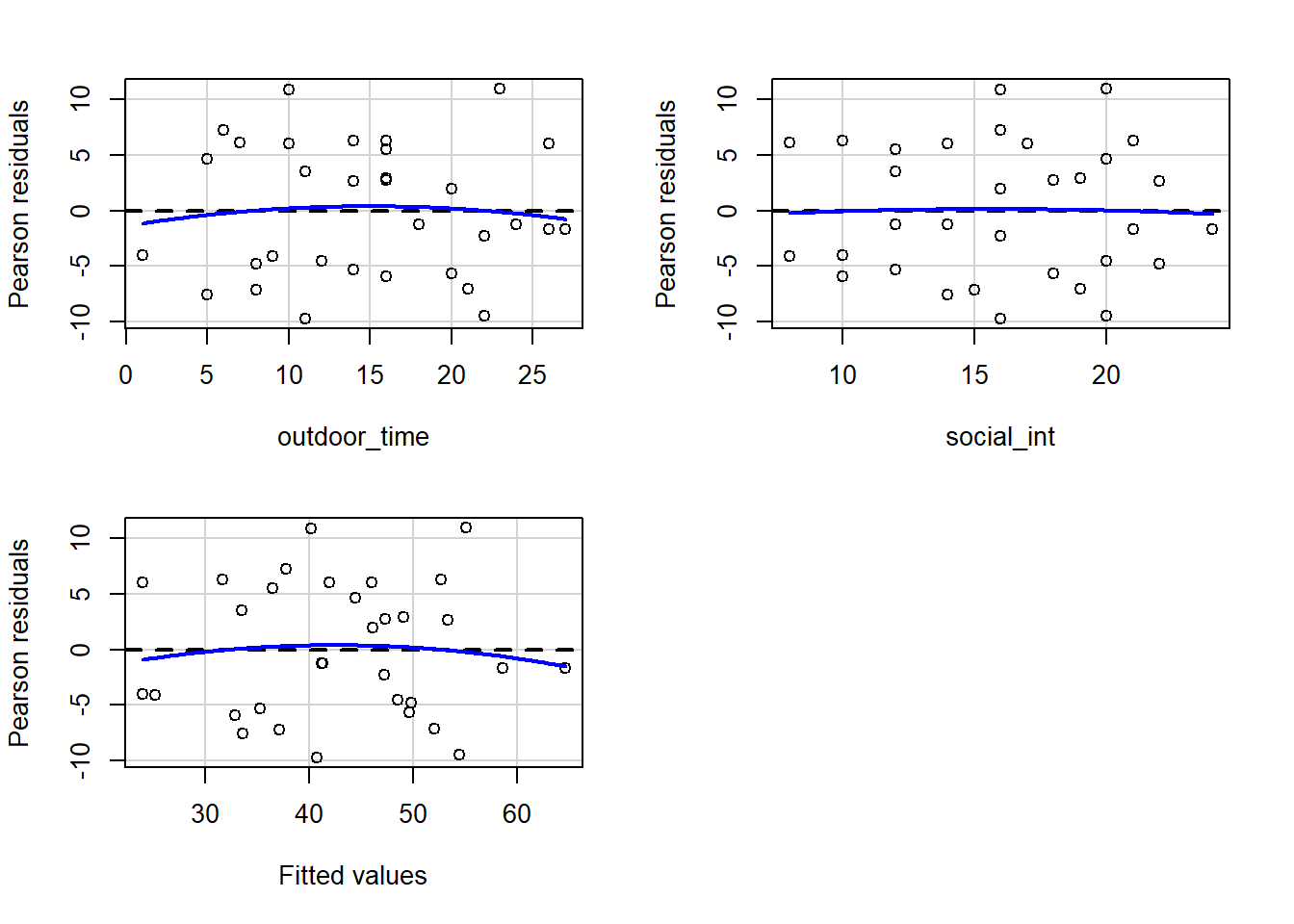
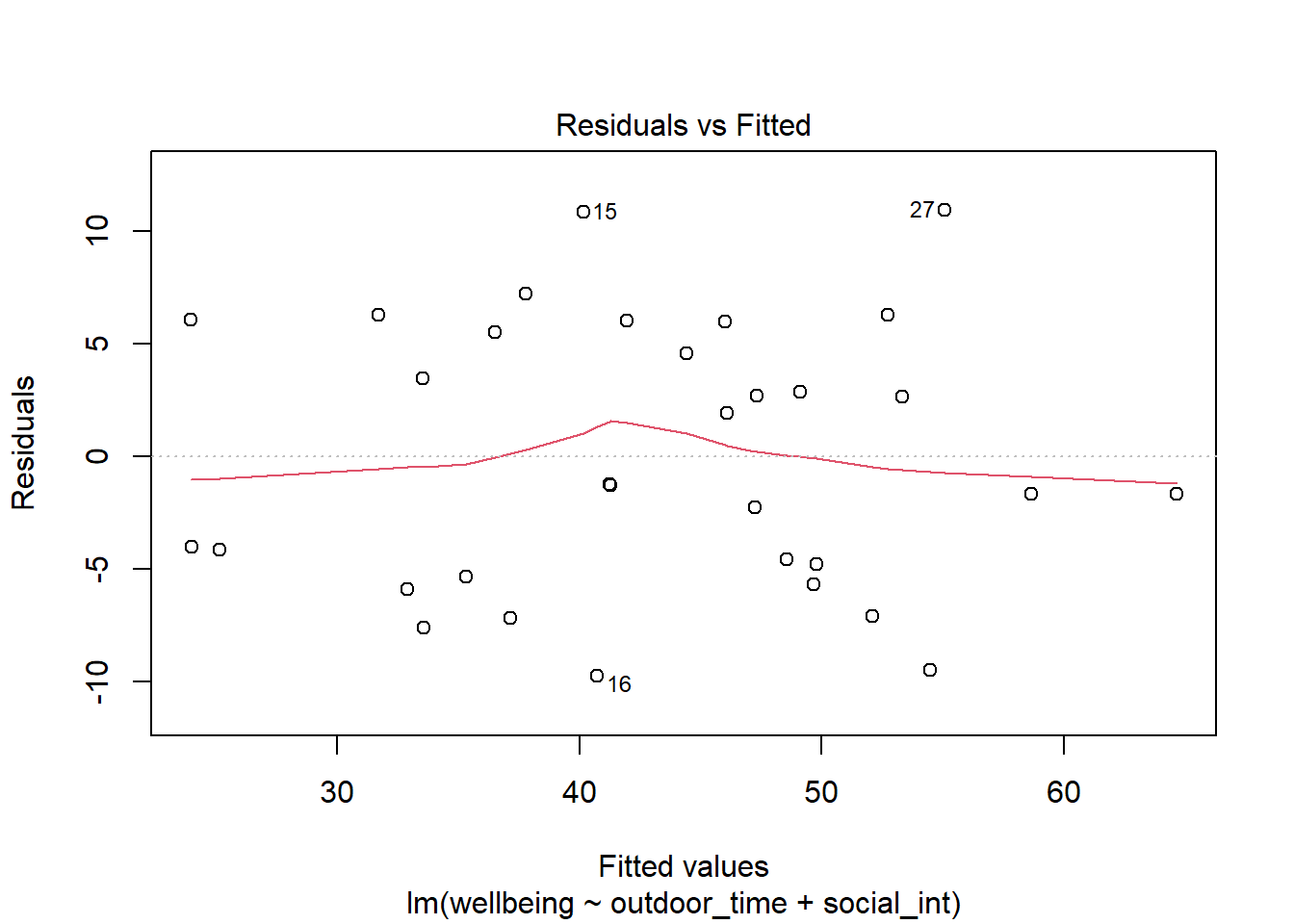
 Or we can make our own:
Or we can make our own: