4. Random Effect Structures | Logistic MLM
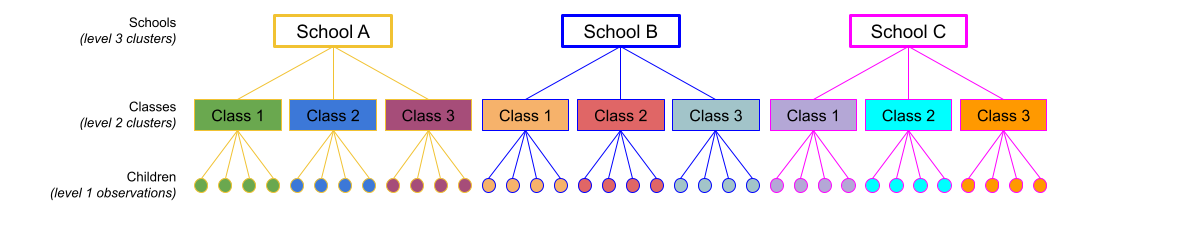
The same principle we have seen for one level of clustering can be extended to clustering at different levels (for instance, observations are clustered within subjects, which are in turn clustered within groups).
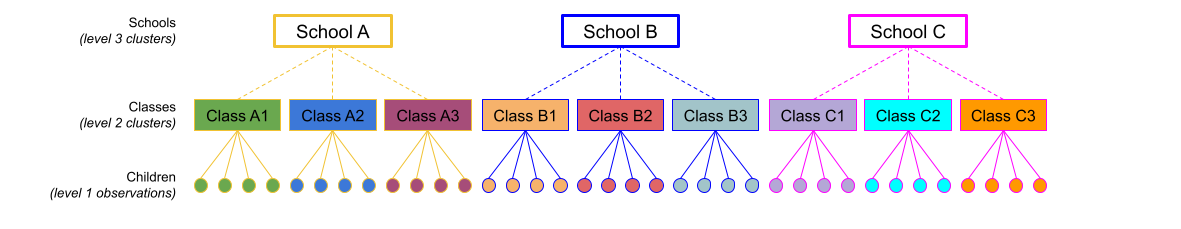
Consider the example where we have observations for each student in every class within a number of schools:
Question: Is “Class 1” in “School 1” the same as “Class 1” in “School 2”?
No.
The classes in one school are distinct from the classes in another even though they are named the same.
The classes-within-schools example is a good case of nested random effects - one factor level (one group in a grouping varible) appears only within a particular level of another grouping variable.
In R, we can specify this using:
(1 | school) + (1 | class:school)
or, more succinctly:
(1 | school/class)
Consider another example, where we administer the same set of tasks for every participant.
Question: Are tasks nested within participants?
No.
Tasks are seen by multiple participants (and participants see multiple tasks).
We could visualise this as the below:
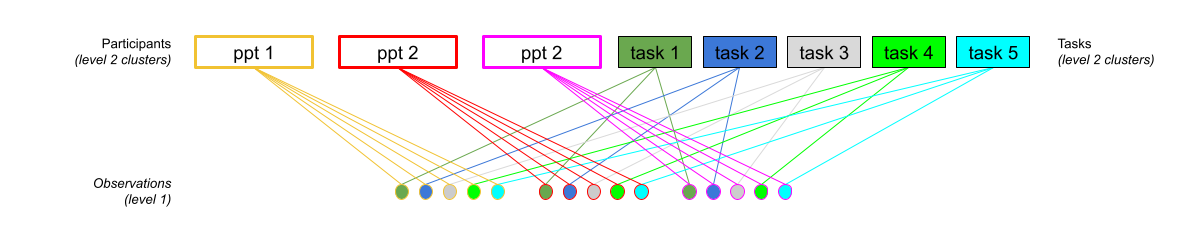
In the sense that these are not nested, they are crossed random effects.
In R, we can specify this using:
(1 | subject) + (1 | task)
Nested vs Crossed
Nested: Each group belongs uniquely to a higher-level group.
Crossed: Not-nested.
Note that in the schools and classes example, had we changed data such that the classes had unique IDs (e.g., see below), then the structures (1 | school) + (1 | class) and (1 | school/class) would give the same results.
Below are a selection of different formulas for specifying different random effect structures, taken from the lme4 vignette. This might look like a lot, but over time and repeated use of multilevel models you will get used to reading these in a similar way to getting used to reading the formula structure of y ~ x1 + x2 in all our linear models.
| Formula | Alternative | Meaning |
|---|---|---|
| \(\text{(1 | g)}\) | \(\text{1 + (1 | g)}\) | Random intercept with fixed mean |
| \(\text{(1 | g1/g2)}\) | \(\text{(1 | g1) + (1 | g1:g2)}\) | Intercept varying among \(g1\), and \(g2\) within \(g1\) |
| \(\text{(1 | g1) + (1 | g2)}\) | \(\text{1 + (1 | g1) + (1 | g2)}\) | Intercept varying among \(g1\) and \(g2\) |
| \(\text{x + (x | g)}\) | \(\text{1 + x + (1 + x | g)}\) | Correlated random intercept and slope |
| \(\text{x + (x || g)}\) | \(\text{1 + x + (1 | g) + (0 + x | g)}\) | Uncorrelated random intercept and slope |
Table 1: Examples of the right-hand-sides of mixed effects model formulas. \(g\), \(g1\), \(g2\) are grouping factors, \(x\) is a predictor variable.
You may have noticed that some of our models over the last few weeks have been giving a warning: boundary (singular) fit: see ?isSingular.
Up to now, we’ve been largely ignoring these warnings. However, this week we’re going to look at how to deal with this issue.
boundary (singular) fit: see ?isSingular
The warning is telling us that our model has resulted in a ‘singular fit’. Singular fits often indicate that the model is ‘overfitted’ - that is, the random effects structure which we have specified is too complex to be supported by the data.
Perhaps the most intuitive advice would be remove the most complex part of the random effects structure (i.e. random slopes). This leads to a simpler model that is not over-fitted. In other words, start simplifying from the top (where the most complexity is) to the bottom (where the lowest complexity is). Additionally, when variance estimates are very low for a specific random effect term, this indicates that the model is not estimating this parameter to differ much between the levels of your grouping variable. It might, in some experimental designs, be perfectly acceptable to remove this or simply include it as a fixed effect.
A key point here is that when fitting a mixed model, we should think about how the data are generated. Asking yourself questions such as “do we have good reason to assume subjects might vary over time, or to assume that they will have different starting points (i.e., different intercepts)?” can help you in specifying your random effect structure
You can read in depth about what this means by reading the help documentation for ?isSingular. For our purposes, a relevant section is copied below:
… intercept-only models, or 2-dimensional random effects such as intercept + slope models, singularity is relatively easy to detect because it leads to random-effect variance estimates of (nearly) zero, or estimates of correlations that are (almost) exactly -1 or 1.
Issues of non-convergence can be caused by many things. If you’re model doesn’t converge, it does not necessarily mean the fit is incorrect, however it is is cause for concern, and should be addressed, else you may end up reporting inferences which do not hold.
There are lots of different things which you could do which might help your model to converge. A select few are detailed below:
double-check the model specification and the data
adjust stopping (convergence) tolerances for the nonlinear optimizer, using the optCtrl argument to [g]lmerControl. (see
?convergencefor convergence controls).- What is “tolerance”? Remember that our optimizer is the the method by which the computer finds the best fitting model, by iteratively assessing and trying to maximise the likelihood (or minimise the loss).
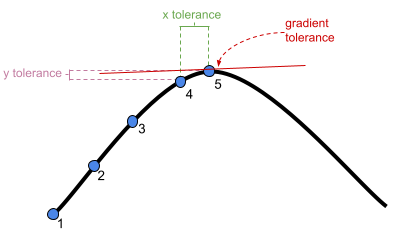
center and scale continuous predictor variables (e.g. with
scale)Change the optimization method (for example, here we change it to
bobyqa):lmer(..., control = lmerControl(optimizer="bobyqa"))
glmer(..., control = glmerControl(optimizer="bobyqa"))Increase the number of optimization steps:
lmer(..., control = lmerControl(optimizer="bobyqa", optCtrl=list(maxfun=50000))
glmer(..., control = glmerControl(optimizer="bobyqa", optCtrl=list(maxfun=50000))Use
allFit()to try the fit with all available optimizers. This will of course be slow, but is considered ‘the gold standard’; “if all optimizers converge to values that are practically equivalent, then we would consider the convergence warnings to be false positives.”Consider simplifying your model, for example by removing random effects with the smallest variance (but be careful to not simplify more than necessary, and ensure that your write up details these changes)
Random effect structures can get pretty complicated quite quickly. Very often it is not the random effects part that is of specific interest to us, but we wish to estimate random effects in order to more accurately partition up the variance in our outcome variable and provide better estimates of fixed effects.
It is a fine balance between fitting the most sophisticated model structure that we possibly can, and fitting a model that converges without too much simplification. Typically for many research designs, the following steps will keep you mostly on track to finding the maximal model:
lmer(outcome ~ fixed effects + (random effects | grouping structure))
Specify the
outcome ~ fixed effectsbit first.- The outcome variable should be clear: it is the variable we are wishing to explain/predict.
- The fixed effects are the things we want to use to explain/predict variation in the outcome variable. These will often be the things that are of specific inferential interest, and other covariates. Just like the simple linear model.
If there is a grouping structure to your data, and those groups (preferably n>7 or 8) are perceived as a random sample of a wider population (the specific groups aren’t interesting to you), then consider including random intercepts (and possibly random slopes of predictors) for those groups
(1 | grouping).If there are multiple different grouping structures, is one nested within another? (If so, we can specify this as
(1 | high_level_grouping / low_level_grouping). If the grouping structures are not nested, we specify them as crossed:(1 | grouping1) + (1 | grouping2).If any of the things in the fixed effects vary within the groups, it might be possible to also include them as random effects.
- as a general rule, don’t specify random effects that are not also specified as fixed effects (an exception could be specifically for model comparison, to isolate the contribution of the fixed effect).
- For things that do not vary within the groups, it rarely makes sense to include them as random effects. For instance if we had a model with
lmer(score ~ genetic_status + (1 + genetic_status | patient))then we would be trying to model a process where “the effect of genetic_status on scores is different for each patient”. But if you consider an individual patient, their genetic status never changes. For patient \(i\), what is “the effect of genetic status on score”? It’s undefined. This is because genetic status only varies between patients. - Sometimes, things will vary within one grouping, but not within another. E.g. with patients nested within hospitals
(1 | hospital/patient), genetic_status varies between patients, but within hospitals. Therefore we could theoretically fit a random effect ofgenetic_status | hospital, but not one forgenetic_status | patient.
We can include these while preserving the nested structure of the data by separating out the groupings:(1 + genetic_status | hospital) + (1 | patient:hospital).
- as a general rule, don’t specify random effects that are not also specified as fixed effects (an exception could be specifically for model comparison, to isolate the contribution of the fixed effect).
A common approach to fitting multilevel models is to fit the most complex model that the study design allows. This means including random effects for everything that makes theoretical sense to be included. Because this “maximal model” will often not converge, we then simplify our random effect structure to obtain a convergent model.
Candidate random effect terms for removal are:
- overly complex terms (e.g.
(1 + x1 * x2 | group)could be simplified to(1 + x1 + x2 | group)).
- those with little variance
- those with near perfect correlation (we can alternatively remove just the correlation)
- Obtain final model, and proceed to checking assumptions, checking influence, plotting, and eventually writing up.
Exercises: Crossed Ranefs
Data: Test-enhanced learning
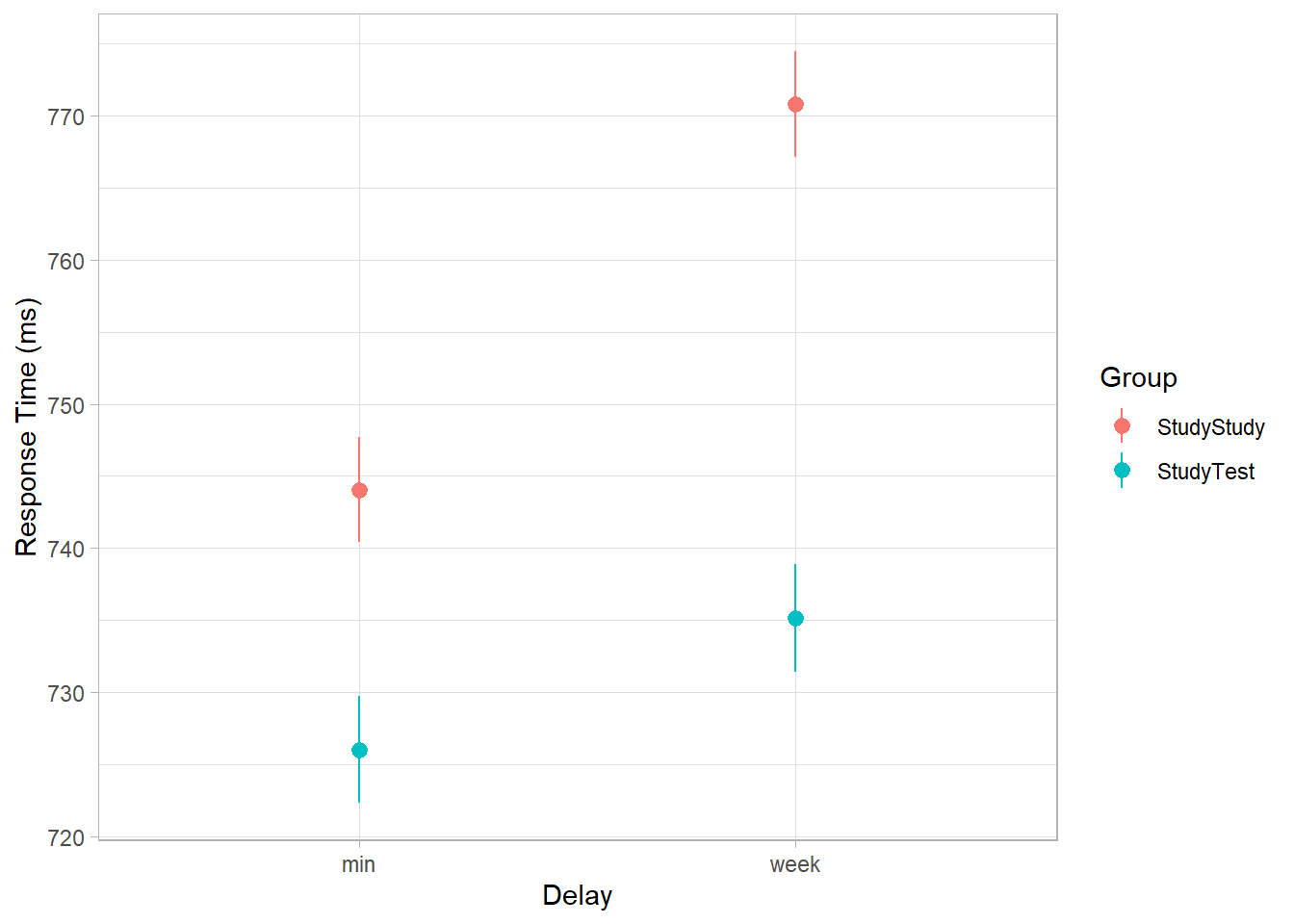
An experiment was run to conceptually replicate “test-enhanced learning” (Roediger & Karpicke, 2006): two groups of 25 participants were presented with material to learn. One group studied the material twice (StudyStudy), the other group studied the material once then did a test (StudyTest). Recall was tested immediately (one minute) after the learning session and one week later. The recall tests were composed of 175 items identified by a keyword (Test_word).
The researchers are interested in how test-enhanced learning influences time-to-recall. The critical (replication) prediction is that the StudyTest group will retain the material better (lower reaction times) on the 1-week follow-up test compared to the StudyStudy group.
| variable | description |
|---|---|
| Subject_ID | Unique Participant Identifier |
| Group | Group denoting whether the participant studied the material twice (StudyStudy), or studied it once then did a test (StudyTest) |
| Delay | Time of recall test ('min' = Immediate, 'week' = One week later) |
| Test_word | Word being recalled (175 different test words) |
| Rtime | Time to respond (milliseconds) |
The following code loads the data into your R environment by creating a variable called tel:
load(url("https://uoepsy.github.io/data/RTtestlearning.RData"))Load and plot the data.
For this week, we’ll use Reaction Time as our proxy for the test performance, so you’ll probably want that variable on the y-axis.
Does it look like the effect was replicated?
Our outcome variable is reaction time, so we’ll want to plot that on the y axis.
As described in the data summary, this is a replication, and the prediction is that the StudyTest group will retain the material better (have lower reaction times) on the 1-week follow-up test compared to the StudyStudy group.
So we want to show the reaction times for each group, at each test (min or week delay). Do we want to show all the datapoints, or maybe just the average for each group at each test (in which case, does stat_summary() help?).
The critical (replication) prediction is that the
StudyStudygroup should perform somewhat better on the immediate recall test, but theStudyTestgroup will retain the material better and thus perform better on the 1-week follow-up test.
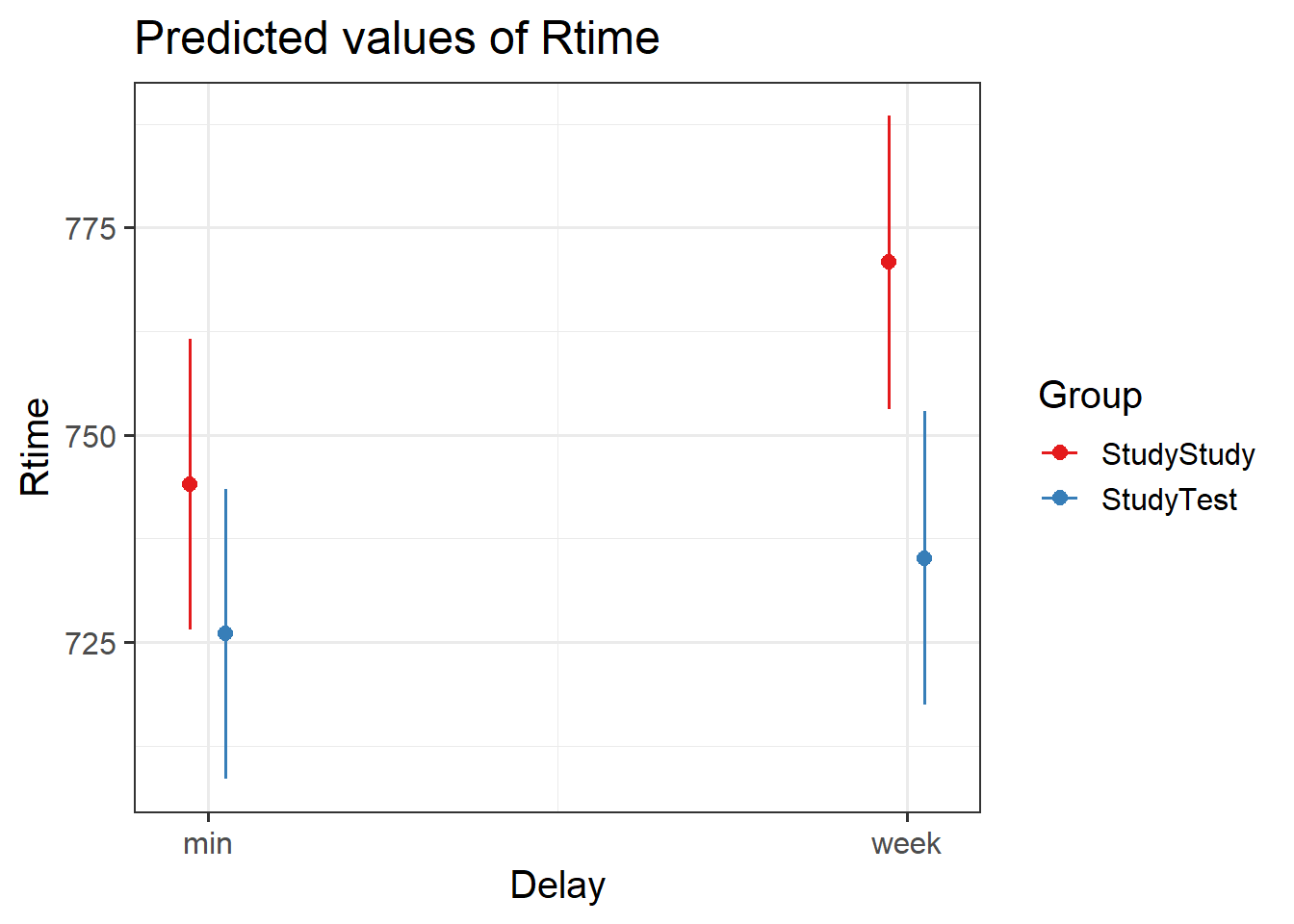
Test the critical hypothesis using a multi-level model.
Try to fit the maximally complex random effect structure that is supported by the experimental design.
NOTE: Your model probably won’t converge. We’ll deal with that in the next question
- We can expect variability across subjects (some people are better at learning than others) and across items (some of the recall items are harder than others). How should this be represented in the random effects?
- If a model takes ages to fit, you might want to cancel it by pressing the escape key. It is normal for complex models to take time, but for the purposes of this task, give up after a couple of minutes, and try simplifying your model.
Often, models with maximal random effect structures will not converge, or will obtain a singular fit. So we need to simplify the model until we achieve convergence (Barr et al., 2013).
Incrementally simplify your model from the previous question until you obtain a model that converges and is not a singular fit.
you can look at the variance estimates and correlations easily by using the VarCorr() function. What jumps out?
There’s a bit of subjectivity here about which simplifications we can live with, so you might end up with a slightly different model from the solutions, and that’s okay.
Exercises: Nested Random Effects
Data: Naming
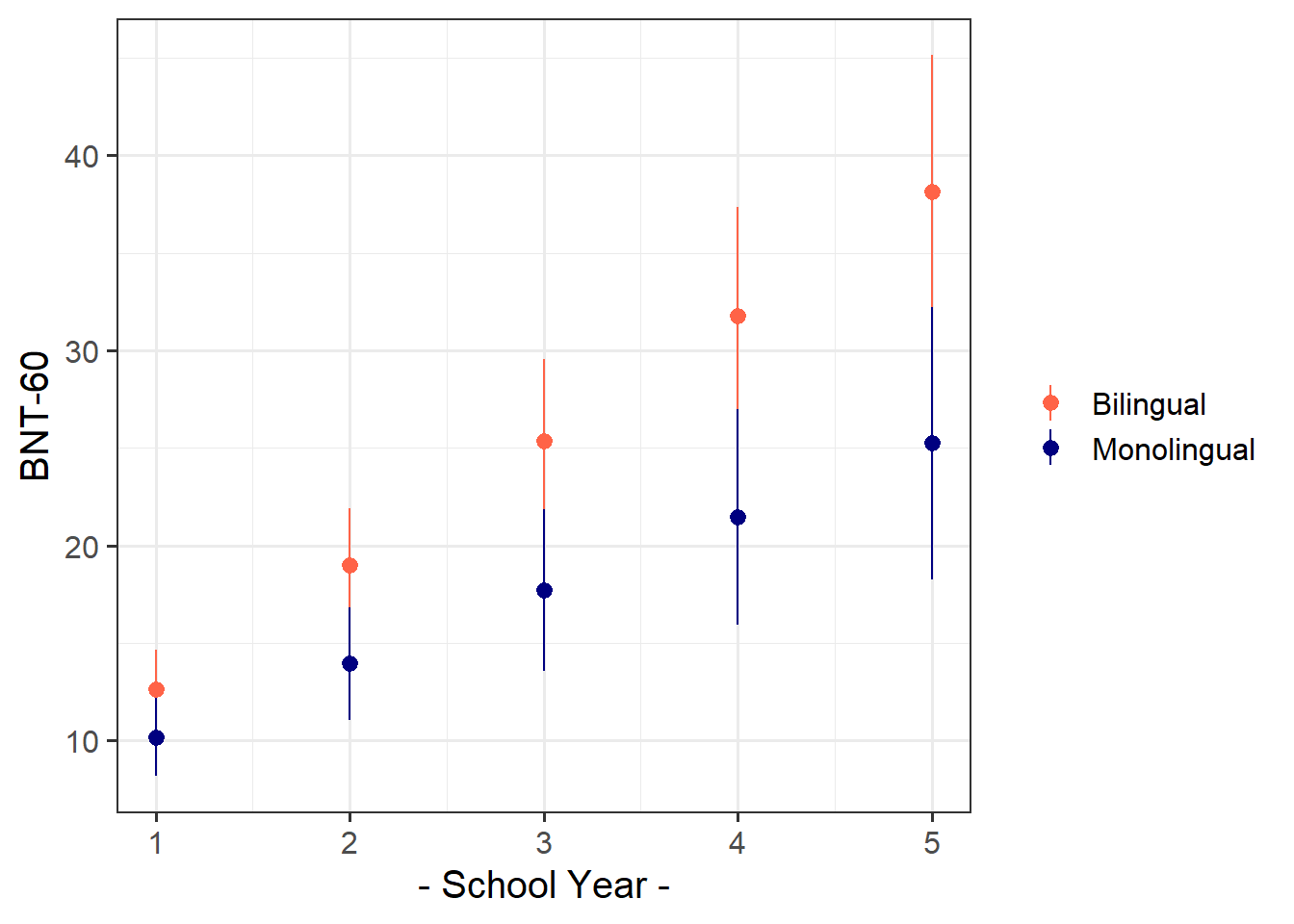
74 children from 10 schools were administered the full Boston Naming Test (BNT-60) on a yearly basis for 5 years to examine development of word retrieval. Five of the schools taught lessons in a bilingual setting with English as one of the languages, and the remaining five schools taught in monolingual English.
The data is available at https://uoepsy.github.io/data/bntmono.csv.
| variable | description |
|---|---|
| child_id | unique child identifier |
| school_id | unique school identifier |
| BNT60 | score on the Boston Naming Test-60. Scores range from 0 to 60 |
| schoolyear | Year of school |
| mlhome | Mono/Bi-lingual School. 0 = Bilingual, 1 = Monolingual |
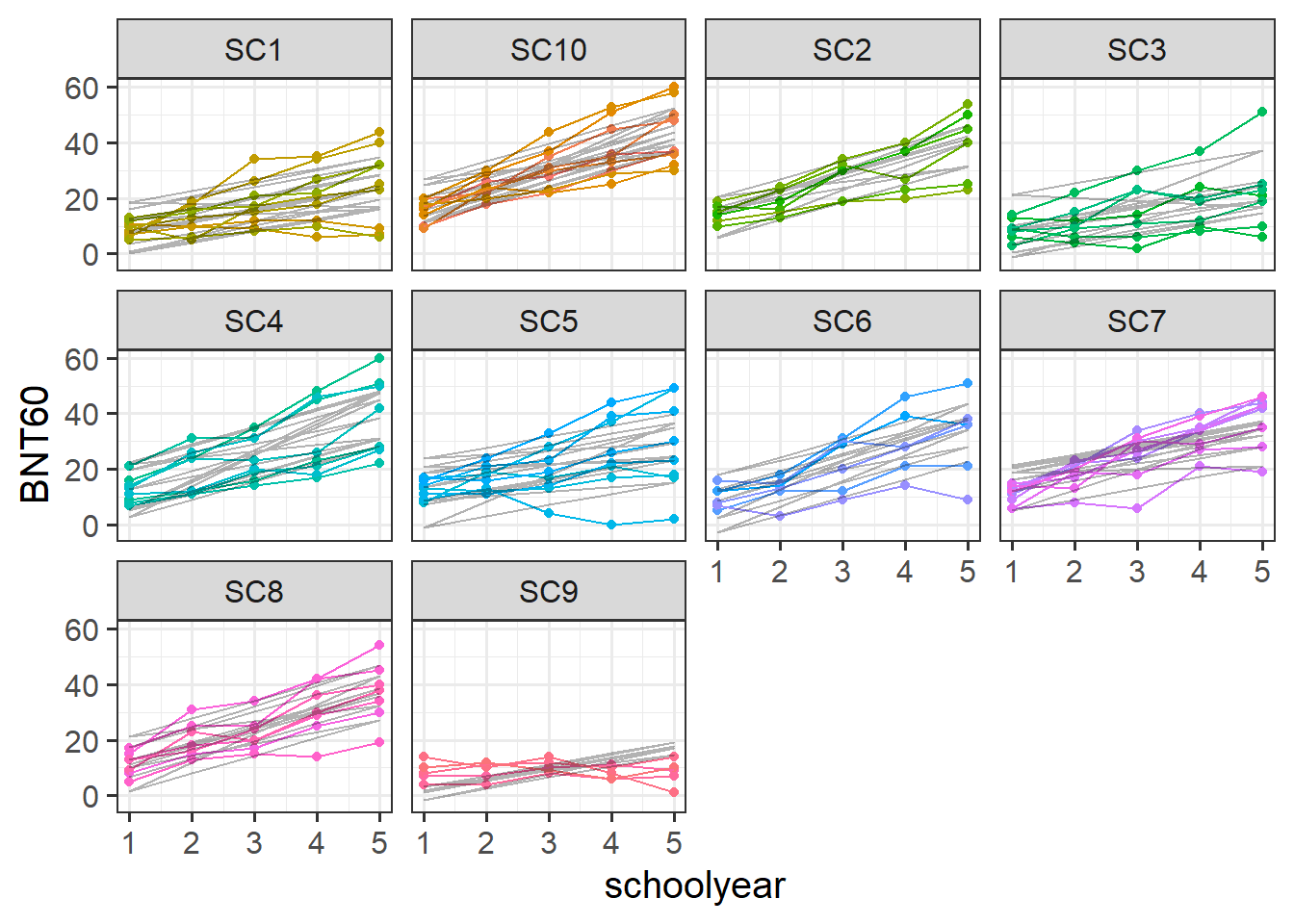
Let’s start by thinking about our clustering - we’d like to know how much of the variance in BNT60 scores is due to the clustering of data within children, who are themselves within schools. One easy way of assessing this is to fit an intercept only model, which has the appropriate random effect structure.
Using the model below, calculate the proportion of variance attributable to the clustering of data within children within schools.
bnt_null <- lmer(BNT60 ~ 1 + (1 | school_id/child_id), data = bnt)the random intercept variances are the building blocks here. There are no predictors in this model, so all the variance in the outcome gets attributed to either school-level nesting, child-level nesting, or else is lumped into the residual.
Fit a model examining the interaction between the effects of school year and mono/bilingual teaching on word retrieval (via BNT test), with random intercepts only for children and schools.
make sure your variables are of the right type first - e.g. numeric, factor etc
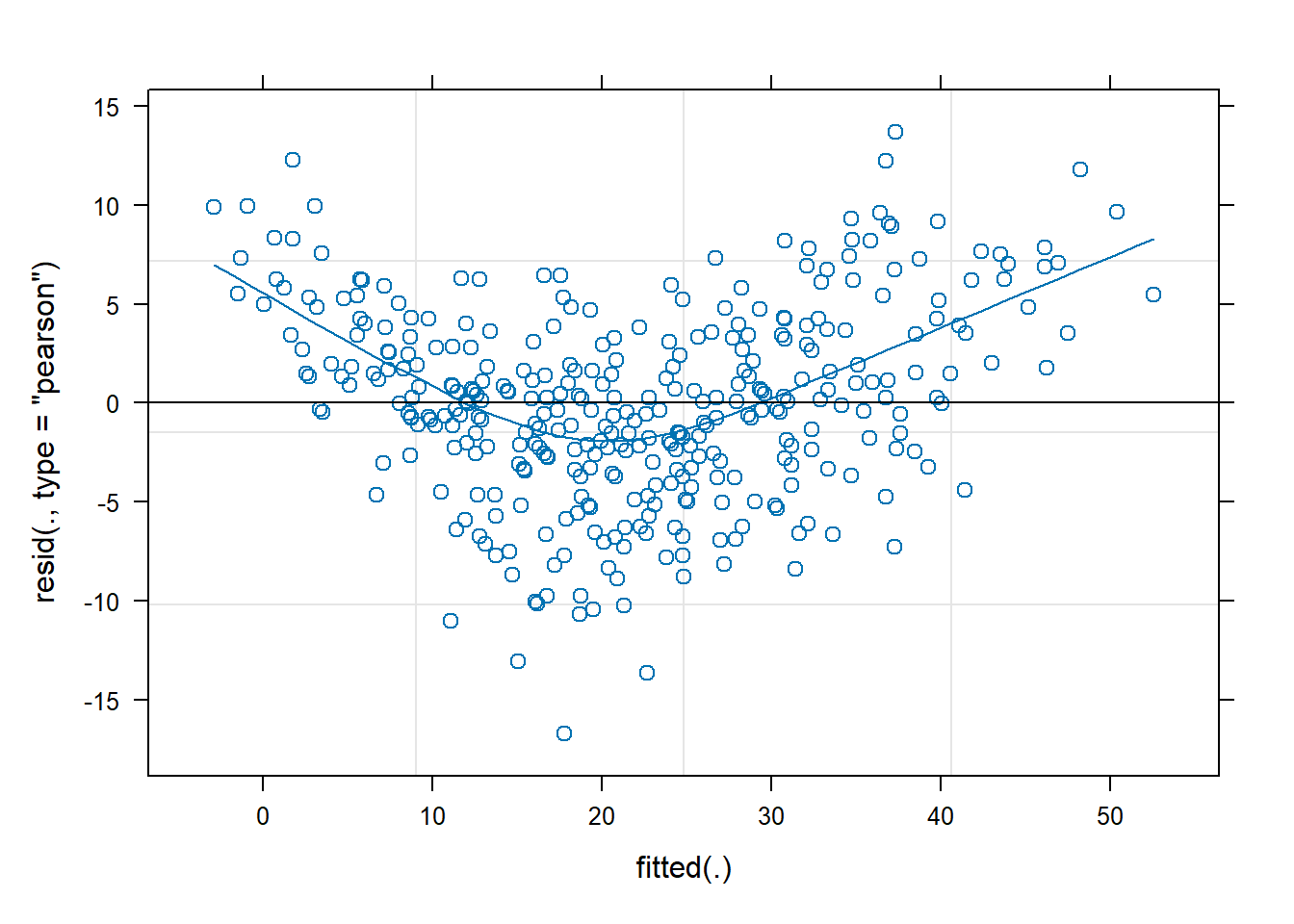
Examine the fit and consider your model assumptions, and assess what might be done to improve the model in order to make better statistical inferences.
Using a method of your choosing, conduct inferences (i.e. obtain p-values or confidence intervals) from your final model and write up the results.
Optional Exercises: Logistic MLM
Don’t forget to look back at other materials!
Back in DAPR2, we introduced logistic regression in semester 2, week 8. The lab contained some simulated data based on a hypothetical study about inattentional blindness. That content will provide a lot of the groundwork for this week, so we recommend revisiting it if you feel like it might be useful.
lmer() to glmer()
Remember how we simply used glm() and could specify the family = "binomial" in order to fit a logistic regression? Well it’s much the same thing for multi-level models!
- Gaussian model:
lmer(y ~ x1 + x2 + (1 | g), data = data)
- Binomial model:
glmer(y ~ x1 + x2 + (1 | g), data = data, family = binomial(link='logit'))- or just
glmer(y ~ x1 + x2 + (1 | g), data = data, family = "binomial") - or
glmer(y ~ x1 + x2 + (1 | g), data = data, family = binomial)
- or just
Data: Memory Recall & Finger Tapping
Research Question: After accounting for effects of sentence length, does the rhythmic tapping of fingers aid memory recall?
Researchers recruited 40 participants. Each participant was tasked with studying and then recalling 10 randomly generated sentences between 1 and 14 words long. For 5 of these sentences, participants were asked to tap their fingers along with speaking the sentence in both the study period and in the recall period. For the remaining 5 sentences, participants were asked to sit still.
The data are available at https://uoepsy.github.io/data/memorytap.csv, and contains information on the length (in words) of each sentence, the condition (static vs tapping) under which it was studied and recalled, and whether the participant was correct in recalling it.
| variable | description |
|---|---|
| ppt | Participant Identifier (n=40) |
| slength | Number of words in sentence |
| condition | Condition under which sentence is studied and recalled ('static' = sitting still, 'tap' = tapping fingers along to sentence) |
| correct | Whether or not the sentence was correctly recalled |
Research Question: After accounting for effects of sentence length, does the rhythmic tapping of fingers aid memory recall?
Fit an appropriate model to answer the research question.
- our outcome is conceptually ‘memory recall’, and it’s been measured by “Whether or not a sentence was correctly recalled”. This is a binary variable.
- we have multiple observations for each ?????
This will define our( | ??? )bit
Take some time to remind yourself from DAPR2 of the interpretation of logistic regression coefficients.
In family = binomial(link='logit'), we are modelling the log-odds. We can obtain estimates on this scale using:
fixef(model)summary(model)$coefficientstidy(model)from broom.mixed
- (there are probably more ways, but I can’t think of them right now!)
We can use exp(), to get these back into odds and odds ratios.
Interpret each of the fixed effect estimates from your model.
Checking the assumptions in non-gaussian models in general (i.e. those where we set the family to some other error distribution) can be a bit tricky, and this is especially true for multilevel models.
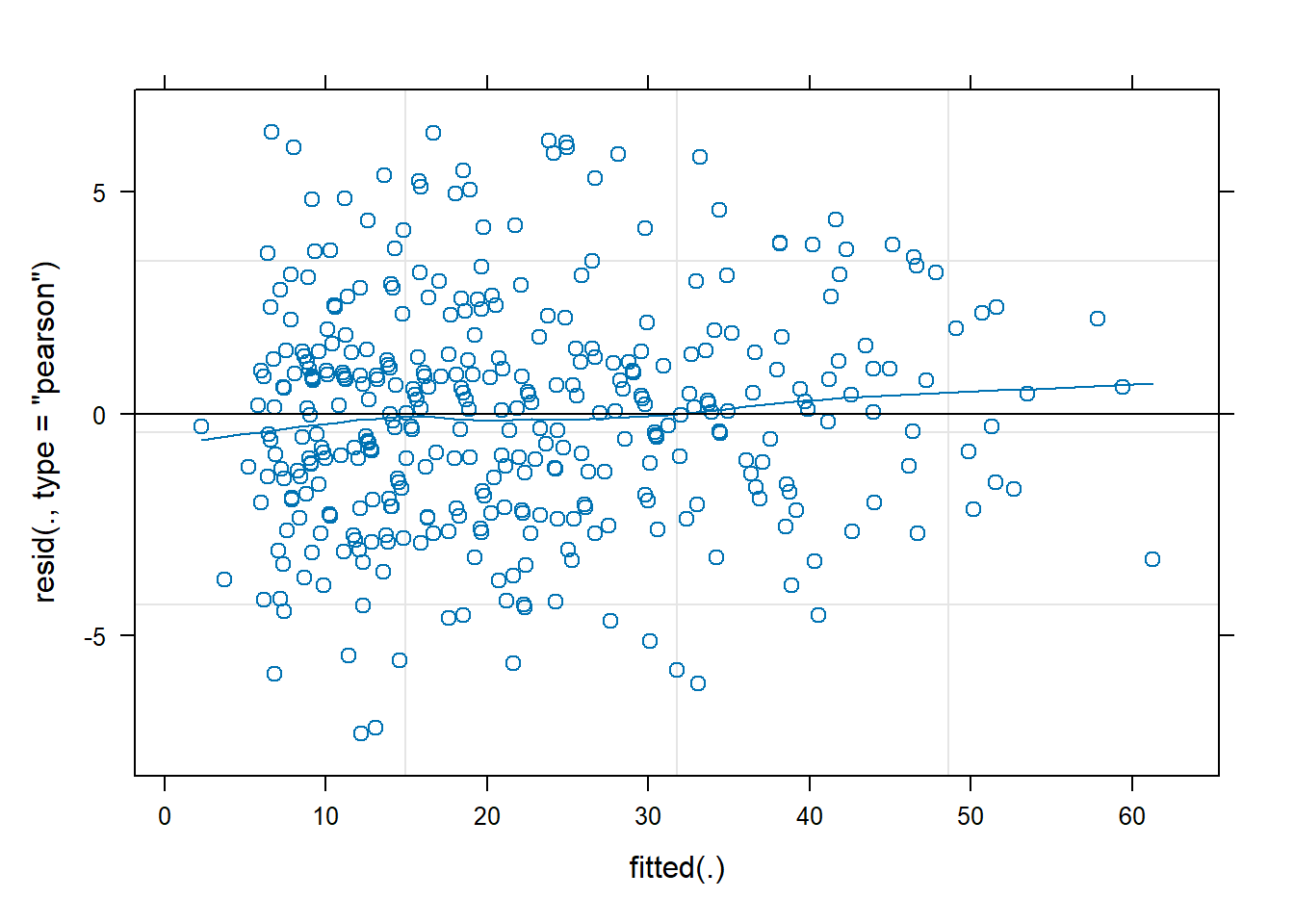
For the logistic MLM, the standard assumptions of normality etc for our Level 1 residuals residuals(model) do not hold. However, it is still useful to quickly plot the residuals and check that \(|residuals|\leq 2\) (or \(|residuals|\leq 3\) if you’re more relaxed). We don’t need to worry too much about the pattern though.
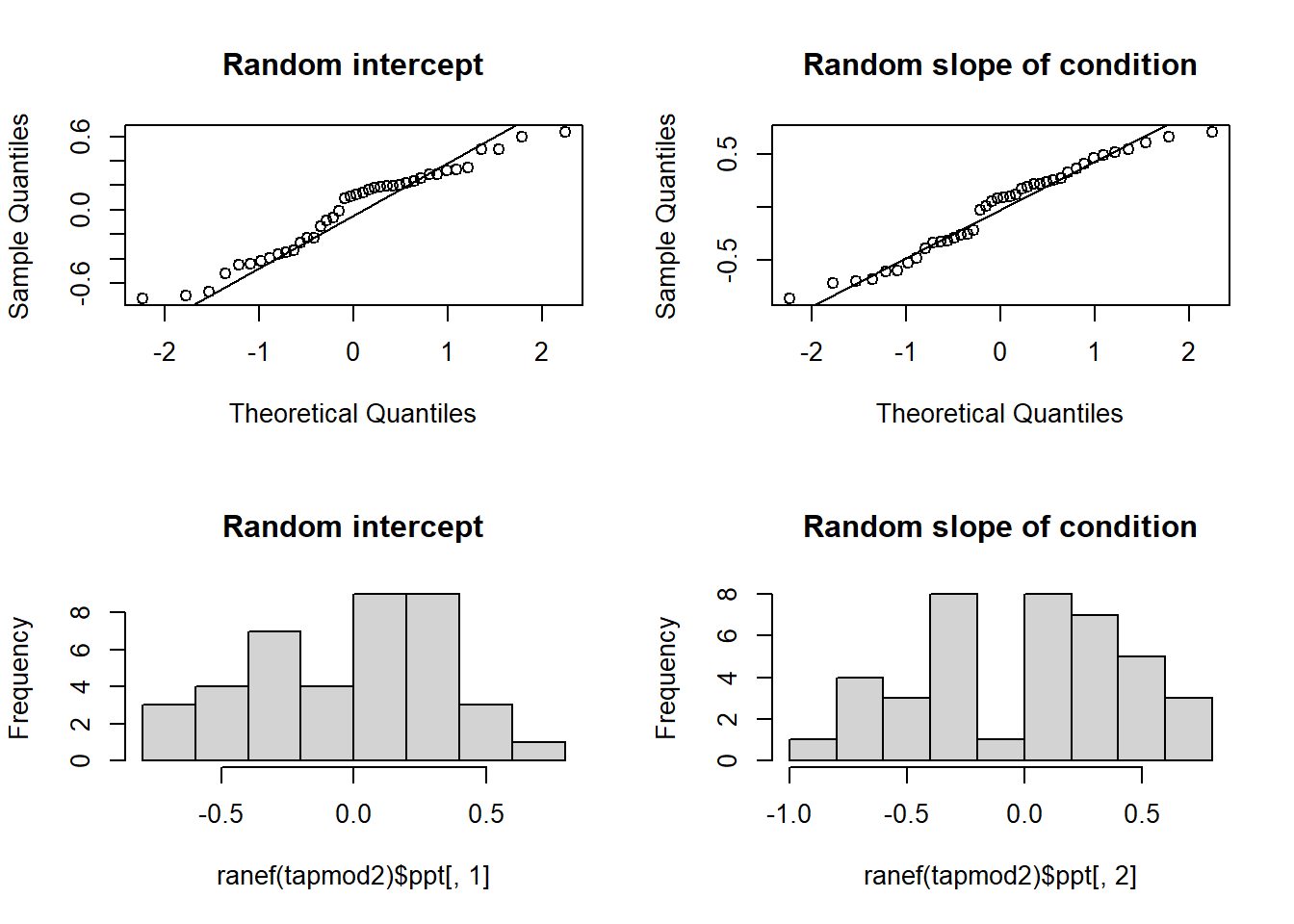
While we’re more relaxed about Level 1 residuals, we do still want our random effects ranef(model) to look fairly normally distributed.
- Plot the level 1 residuals and check whether any are greater than 3 in magnitude
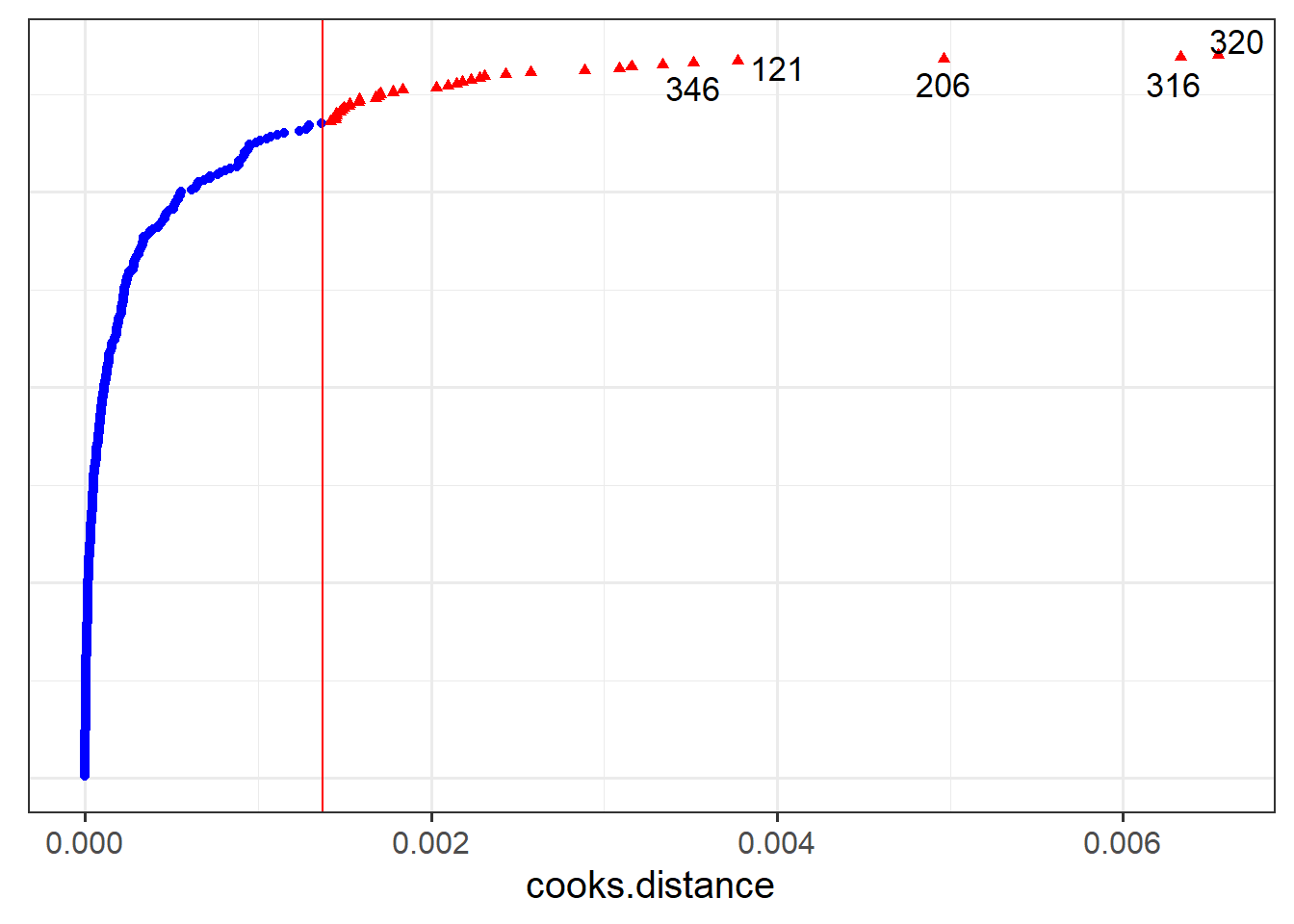
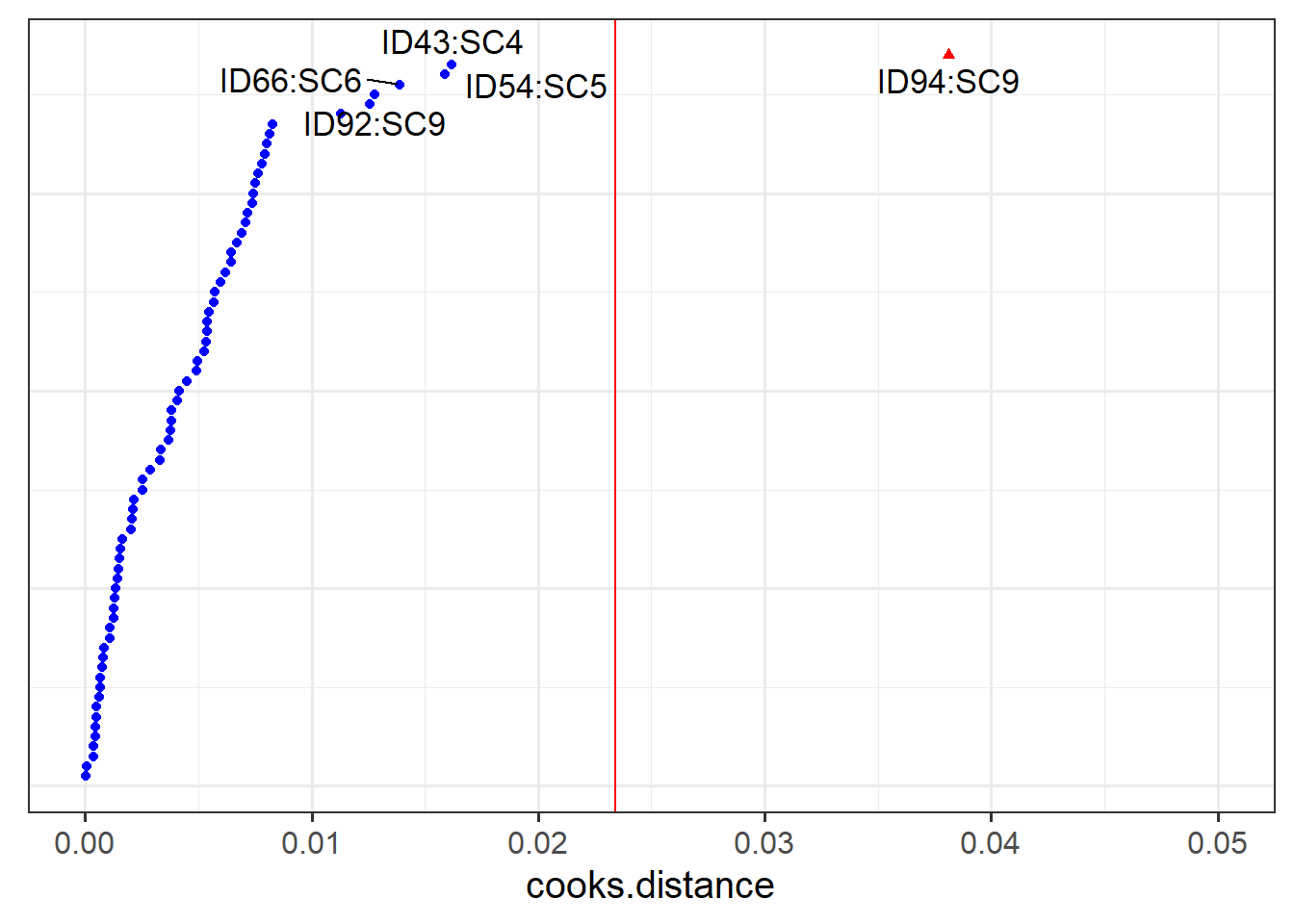
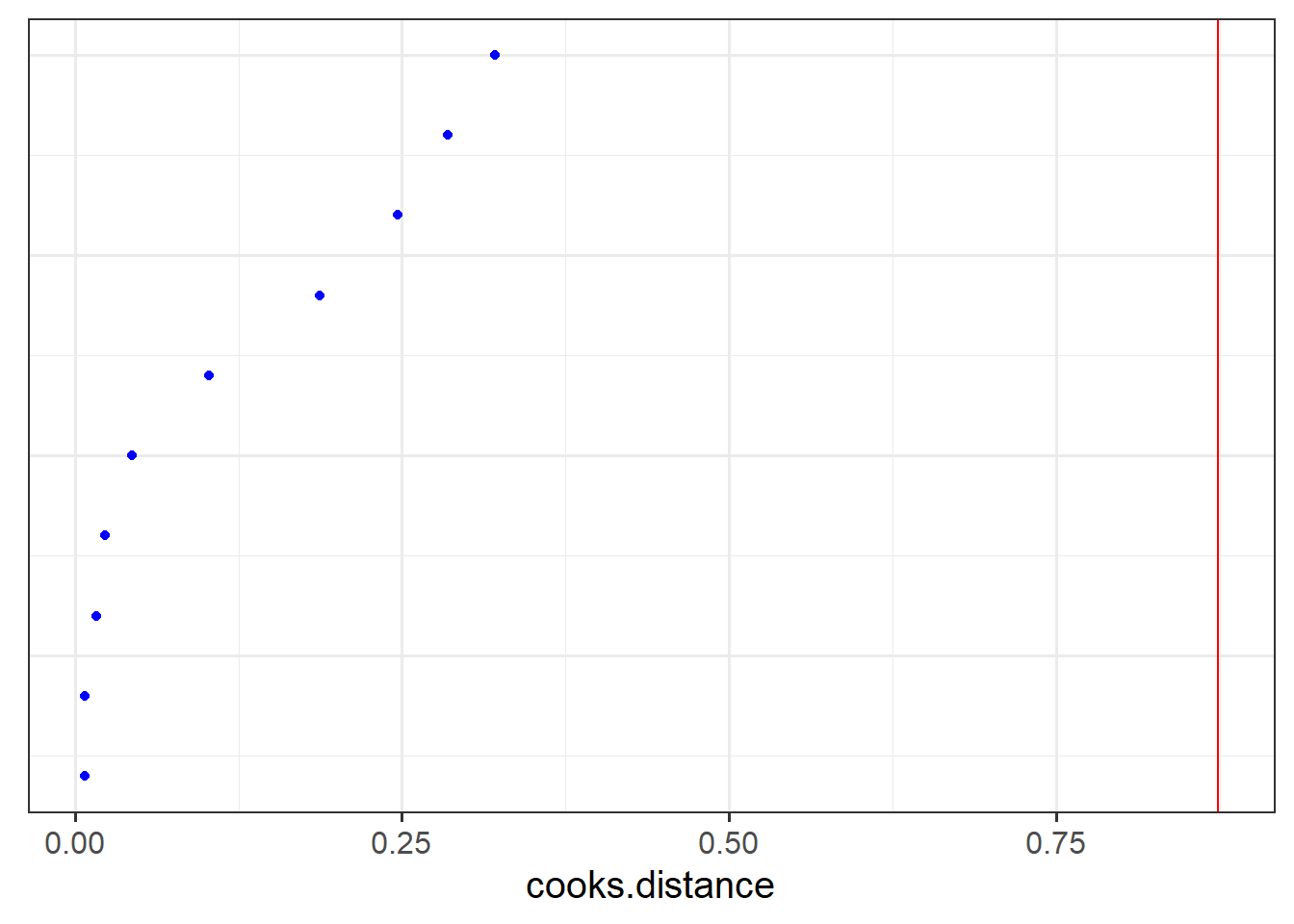
- Plot the random effects (the level 2 residuals) and assess the normality.
- The HLMdiag package doesn’t support diagnosing influential points/clusters for
glmer, but there is a package called influence.me which might help: https://journal.r-project.org/archive/2012/RJ-2012-011/RJ-2012-011.pdf - There are packages which aim to create more interpretable residual plots for these models via simulation, such as the DHARMa package: https://cran.r-project.org/web/packages/DHARMa/vignettes/DHARMa.html
Practice Datasets Weeks 4 and 5
Below are various datasets on which you can try out your new-found modelling skills. Read the descriptions carefully, keeping in mind the explanation of how the data is collected and the research question that motivates the study design.
These data are simulated to represent data from a fake experiment, in which participants were asked to drive around a route in a 30mph zone. Each participant completed the route 3 times (i.e. “repeated measures”), but each time they were listening to different audio (either speech, classical music or rap music). Their average speed across the route was recorded. This is a fairly simple design, that we might use to ask “how is the type of audio being listened to associated with driving speeds?”
The data are available at https://uoepsy.github.io/data/drivingmusicwithin.csv.
| variable | description |
|---|---|
| pid | Participant Identifier |
| speed | Avg Speed Driven on Route (mph) |
| music | Music listened to while driving (classical music / rap music / spoken word) |
These data are simulated to represent data from 50 participants, each measured at 3 different time-points (pre, during, and post) on a measure of stress. Participants were randomly allocated such that half received some cognitive behavioural therapy (CBT) treatment, and half did not. This study is interested in assessing whether the two groups (control vs treatment) differ in how stress changes across the 3 time points.
The data are available at https://uoepsy.github.io/data/stressint.csv.
| variable | description |
|---|---|
| ppt | Participant Identifier |
| stress | Stress (range 0 to 100) |
| time | Time (pre/post/during) |
| group | Whether participant is in the CBT group or control group |
These data are simulated to represent data from 30 participants who took part in an experiment designed to investigate whether fluency of speech influences how believable an utterance is perceived to be.
Each participant listened to the same 20 statements, with 10 being presented in fluent speech, and 10 being presented with a disfluency (an “erm, …”). Fluency of the statements was counterbalanced such that 15 participants heard statements 1 to 10 as fluent and 11 to 20 as disfluent, and the remaining 15 participants heard statements 1 to 10 as disfluent, and 11 to 20 as fluent. The order of the statements presented to each participant was random. Participants rated each statement on how believable it is on a scale of 0 to 100.
The data are available at https://uoepsy.github.io/data/erm_belief.csv.
| variable | description |
|---|---|
| ppt | Participant Identifier |
| trial_n | Trial number |
| sentence | Statement identifier |
| condition | Condition (fluent v disfluent) |
| belief | belief rating (0-100) |
| statement | Statement |
These data are simulated to represent a large scale international study of cognitive aging, for which data from 17 research centers has been combined. The study team are interested in whether different cognitive domains have different trajectories as people age. Do all cognitive domains decline at the same rate? Do some decline more steeply, and some less? The literature suggests that scores on cognitive ability are predicted by educational attainment, so they would like to control for this.
Each of the 17 research centers recruited a minimum of 14 participants (Median = 21, Range 14-29) at age 45, and recorded their level of education (in years). Participants were then tested on 5 cognitive domains: processing speed, spatial visualisation, memory, reasoning, and vocabulary. Participants were contacted for follow-up on a further 9 occasions (resulting in 10 datapoints for each participant), and at every follow-up they were tested on the same 5 cognitive domains. Follow-ups were on average 3 years apart (Mean = 3, SD = 0.8).
The data are available at https://uoepsy.github.io/data/cogdecline.csv.
| variable | description |
|---|---|
| cID | Center ID |
| pptID | Participant Identifier |
| educ | Educational attainment (years of education) |
| age | Age at visit (years) |
| processing_speed | Score on Processing Speed domain task |
| spatial_visualisation | Score on Spatial Visualisation domain task |
| memory | Score on Memory domain task |
| reasoning | Score on Reasoning domain task |
| vocabulary | Score on Vocabulary domain task |
Footnotes
It’s always going to be debateable about what is ‘too high’ because in certain situations you might expect correlations close to 1. It’s best to think through whether it is a feasible value given the study itself↩︎