2. Intro to Multilevel Models
The methods we’re going to learn about in the first five weeks of this course are known by lots of different names: “multilevel models”; “hierarchical linear models”; “mixed-effect models”; “mixed models”; “nested data models”; “random coefficient models”; “random-effects models”; “random parameter models”… and so on).
What the idea boils down to is that model parameters vary at more than one level. This week, we’re going to explore what that means.
Throughout this course, we will tend to use the terms “mixed effect model”, “linear mixed model (LMM)” and “multilevel model (MLM)” interchangeably.
Multilevel Models (MLMs) (or “Linear Mixed Models” (LMMs)) take the approach of allowing the groups/clusters to vary around our \(\beta\) estimates.
In the lectures, we saw this as:
\[ \begin{align} & \text{for observation }j\text{ in group }i \\ \quad \\ & \text{Level 1:} \\ & \color{red}{y_{ij}}\color{black} = \color{blue}{\beta_{0i} \cdot 1 + \beta_{1i} \cdot x_{ij}}\color{black} + \varepsilon_{ij} \\ & \text{Level 2:} \\ & \color{blue}{\beta_{0i}}\color{black} = \gamma_{00} + \color{orange}{\zeta_{0i}} \\ & \color{blue}{\beta_{1i}}\color{black} = \gamma_{10} + \color{orange}{\zeta_{1i}} \\ \quad \\ & \text{Where:} \\ & \gamma_{00}\text{ is the population intercept, and }\color{orange}{\zeta_{0i}}\color{black}\text{ is the deviation of group }i\text{ from }\gamma_{00} \\ & \gamma_{10}\text{ is the population slope, and }\color{orange}{\zeta_{1i}}\color{black}\text{ is the deviation of group }i\text{ from }\gamma_{10} \\ \end{align} \]
We are now assuming \(\color{orange}{\zeta_0}\), \(\color{orange}{\zeta_1}\), and \(\varepsilon\) to be normally distributed with a mean of 0, and we denote their variances as \(\sigma_{\color{orange}{\zeta_0}}^2\), \(\sigma_{\color{orange}{\zeta_1}}^2\), \(\sigma_\varepsilon^2\) respectively.
The \(\color{orange}{\zeta}\) components also get termed the “random effects” part of the model, Hence names like “random effects model”, etc.
Many people use the symbol \(u\) in place of \(\zeta\), and in various resources, you are likely to see \(\alpha\) used to denote the intercept instead of \(\beta_0\).
Sometimes, you will see the levels collapsed into one equation, as it might make for more intuitive reading.
This often fits with the name “mixed effects” for these models, as the “effect” of a predictor is a mix of both a fixed and a random part:
\[\color{red}{y_{ij}} = \underbrace{(\gamma_{00} + \color{orange}{\zeta_{0i}})}_{\color{blue}{\beta_{0i}}} \cdot 1 + \underbrace{(\gamma_{10} + \color{orange}{\zeta_{1i}})}_{\color{blue}{\beta_{1i}}} \cdot x_{ij} + \varepsilon_{ij} \\\]
In words, this equation is denoting: \[ \begin{align} \color{red}{\text{outcome}}\color{black} = &(\color{blue}{\text{overall intercept}}\color{black} + \color{orange}{\text{random adjustment to intercept by group}}\color{black}) \cdot 1 \, + \\ &( \color{blue}{\text{overall slope}}\color{black} + \color{orange}{\text{random adjustment to slope by group}} \color{black}) \cdot \text{predictor}\, + \\ & \text{residual} \\ \end{align} \]
To fit multilevel models, we’re going to use the lme4 package, and specifically the functions lmer() and glmer().1
data = dataframe,
REML = logical,
control = lmerControl(options)
)
We write the first bit of our formula just the same as our old friend the normal linear model y ~ 1 + x + x2 + ..., where y is the name of our outcome variable, 1 is the intercept (which we don’t have to explicitly state as it will be included anyway) and x, x2 etc are the names of our explanatory variables.
With lme4, we now have the addition of random effect terms, specified in parenthesis with the | operator (the vertical line | is often found to the left of the z key on QWERTY keyboards). We use the | operator to separate the parameters (intercept, slope etc.) on the LHS, from the grouping variable(s) on the RHS, by which we would like to model these parameters as varying.
Random Intercepts
Let us suppose that we wish to model our intercept not as a fixed constant, but as varying randomly according to some grouping around a fixed center. We can such a model by allowing the intercept to vary by our grouping variable (g below):
lmer(y ~ 1 + x + (1|g), data = df)
\[ \begin{align} & \text{Level 1:} \\ & \color{red}{Y_{ij}} = \color{blue}{\beta_{0i} \cdot 1 + \beta_{1} \cdot X_{ij}} + \varepsilon_{ij} \\ & \text{Level 2:} \\ & \color{blue}{\beta_{0i}} = \gamma_{00} + \color{orange}{\zeta_{0i}} \\ \end{align} \]
Random Intercepts and Slopes
By extension we can also allow the effect y~x to vary between groups, by including the x on the left hand side of | in the random effects part of the call to lmer().
lmer(y ~ 1 + x + (1 + x |g), data = df)
\[ \begin{align} & \text{Level 1:} \\ & \color{red}{y_{ij}} = \color{blue}{\beta_{0i} \cdot 1 + \beta_{1i} \cdot x_{ij}} + \varepsilon_{ij} \\ & \text{Level 2:} \\ & \color{blue}{\beta_{0i}} = \gamma_{00} + \color{orange}{\zeta_{0i}} \\ & \color{blue}{\beta_{1i}} = \gamma_{10} + \color{orange}{\zeta_{1i}} \\ \end{align} \]
We can choose whether to estimate our model parameters with ML (maximum likelihood) or REML (restricted maximum likelihood) with the REML argument of lmer():
data = dataframe,
REML = logical,
control = lmerControl(options)
)
lmer() models are by default fitted with REML, which tends to be better for small samples.
Maximum Likelihood (ML)
Remember back to DAPR2 when we introduced logistic regression, and we briefly discussed Maximum likelihood estimation in an explanation of how models are fitted.
The key idea of maximum likelihood estimation (MLE) is that we (well, the computer) iteratively finds the set of estimates for our model which it considers to best reproduce our observed data. Recall our simple linear regression model of how time spent outdoors (hrs per week) is associated with mental wellbeing: \[ \color{red}{Wellbeing_i} = \color{blue}{\beta_0 \cdot{} 1 + \beta_1 \cdot{} OutdoorTime_{i}} + \varepsilon_i \] There are values of \(\beta_0\) and \(\beta_1\) and \(\sigma_\varepsilon\) which maximise the probability of observing the data that we have. For linear regression, these we obtained these same values a different way, via minimising the sums of squares. This approach is not possible for more complex models (e.g., logistic) which is why we turn to MLE.
To read about the subtle difference between “likelihood” and “probability”, you can find a short explanation here
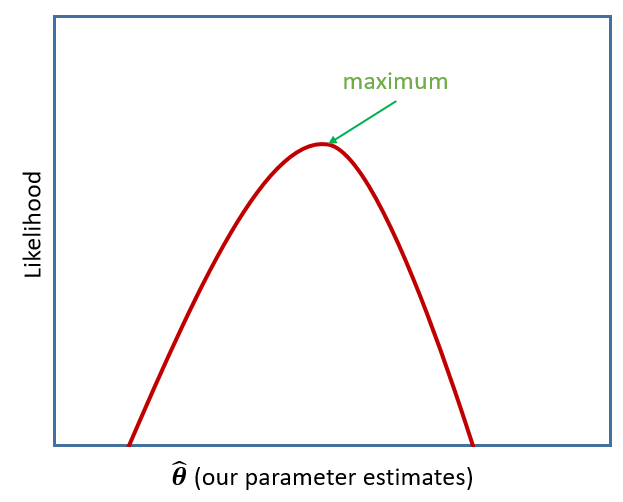
If we are estimating just one single parameter (e.g. a mean), then we can imagine the process of maximum likelihood estimation in a one-dimensional world - simply finding the top of the curve:
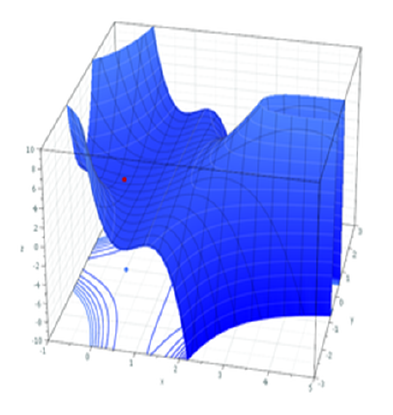
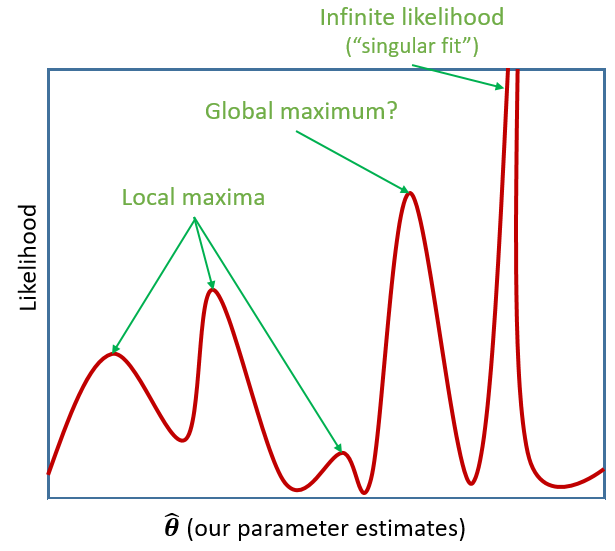
However, our typical models estimate a whole bunch of parameters. The simple regression model above is already having to estimate \(\beta_0\), \(\beta_1\) and \(\sigma_\varepsilon\), and our multi-level models have far more! With lots of parameters being estimated and all interacting to influence the likelihood, our nice curved line becomes a complex surface (see Left panel of Figure 2). So what we (our computers) need to do is find the maximum, but avoid local maxima and singularities (see Figure 3).
Restricted Maximum Likelihood (REML)
When it comes to estimating multilevel models, maximum likelihood will consider the fixed effects as fixed, known values when it estimates the variance components (the random effect variances). This leads to biased estimates of the variance components, specifically biasing them toward being too small, especially if \(n_\textrm{clusters} - n_\textrm{level 2 predictors} - 1 < 50\). This leads to the standard errors of the fixed effects being too small, thereby inflating our type 1 error rate (i.e. greater chance of incorrectly rejecting our null hypothesis).
Restricted Maximum Likelihood (REML) is a method that separates the estimation of fixed and random parts of the model, leading to unbiased estimates of the variance components.
Alongside the ML/REML choice for model estimation, we have some control over the underlying algorithm that is used to move around/search the likelihood surface for our estimates. We’ll learn more about this next week.
data = dataframe,
REML = logical,
control = lmerControl(options)
)
For large datasets and/or complex models (lots of random-effects terms), it is quite common to get a convergence warning. There are lots of different ways to deal with these (to try to rule out hypotheses about what is causing them).
For the time being, if lmer() gives you convergence errors, you could try changing the optimizer. Bobyqa is a good one: add control = lmerControl(optimizer = "bobyqa") when you run your model.
lmer(y ~ 1 + x1 + ... + (1 + .... | g), data = df,
control = lmerControl(optimizer = "bobyqa"))What is a convergence warning??
There are different techniques for maximum likelihood estimation, which we apply by using different ‘optimisers’. Technical problems to do with model convergence and ‘singular fit’ come into play when the optimiser we are using either can’t find a suitable maximum, or gets stuck in a singularity (think of it like a black hole of likelihood, which signifies that there is not enough variation in our data to construct such a complex model).
Exercises: Cross-Sectional
Data: Wellbeing Across Scotland
Recall our dataset from last week, in which we used linear regression to determine how outdoor time (hours per week) is associated with wellbeing in different local authority areas (LAAs) of Scotland. We have data from various LAAs, from Glasgow City, to the Highlands.
scotmw <- read_csv("https://uoepsy.github.io/data/LAAwellbeing.csv")| variable | description |
|---|---|
| ppt | Participant ID |
| name | Participant Name |
| laa | Local Authority Area |
| outdoor_time | Self report estimated number of hours per week spent outdoors |
| wellbeing | Warwick-Edinburgh Mental Wellbeing Scale (WEMWBS), a self-report measure of mental health and well-being. The scale is scored by summing responses to each item, with items answered on a 1 to 5 Likert scale. The minimum scale score is 14 and the maximum is 70. |
| density | LAA Population Density (people per square km) |
Using lmer() from the lme4 package, fit a model predict wellbeing from outdoor_time, with by-LAA random intercepts.
Pass the model to summary() to see the output.
Sometimes the easiest way to start understanding your model is to visualise it.
Load the package broom.mixed. Along with some handy functions tidy() and glance() which give us the information we see in summary(), there is a handy function called augment() which returns us the data in the model plus the fitted values, residuals, hat values, Cook’s D etc..
the broom.mixed package has some useful functions (just like those in the broom package for lm()):
library(broom.mixed)
glance(model) # for overall model stats# A tibble: 1 × 7
nobs sigma logLik AIC BIC REMLcrit df.residual
<int> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 132 5.05 -433. 875. 886. 867. 128tidy(model) # for parameter stats# A tibble: 4 × 6
effect group term estimate std.error statistic
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 fixed <NA> (Intercept) 38.2 2.65 14.4
2 fixed <NA> outdoor_time 0.213 0.0724 2.95
3 ran_pars laa sd__(Intercept) 10.3 NA NA
4 ran_pars Residual sd__Observation 5.05 NA NA augment(model) # observation level stuff (data + model)# A tibble: 132 × 14
wellbeing outdoor_time laa .fitted .resid .hat .cooksd .fixed .mu
<dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 37 20 West Lothian 32.4 4.64 0.139 7.91e-2 42.5 32.4
2 34 23 Falkirk 31.7 2.28 0.192 3.00e-2 43.1 31.7
3 39 29 Falkirk 33.0 6.00 0.195 2.13e-1 44.4 33.0
4 42 21 Scottish Bo… 40.1 1.95 0.163 1.74e-2 42.7 40.1
5 37 10 Dumfries an… 37.4 -0.407 0.167 7.84e-4 40.3 37.4
6 42 19 Argyll and … 43.9 -1.91 0.122 1.12e-2 42.2 43.9
7 38 13 Perth and K… 46.1 -8.06 0.139 2.39e-1 41.0 46.1
8 44 21 East Renfre… 44.4 -0.414 0.168 8.17e-4 42.7 44.4
9 47 16 Inverclyde 42.7 4.30 0.193 1.07e-1 41.6 42.7
10 35 12 Midlothian 33.1 1.94 0.161 1.69e-2 40.8 33.1
# ℹ 122 more rows
# ℹ 5 more variables: .offset <dbl>, .sqrtXwt <dbl>, .sqrtrwt <dbl>,
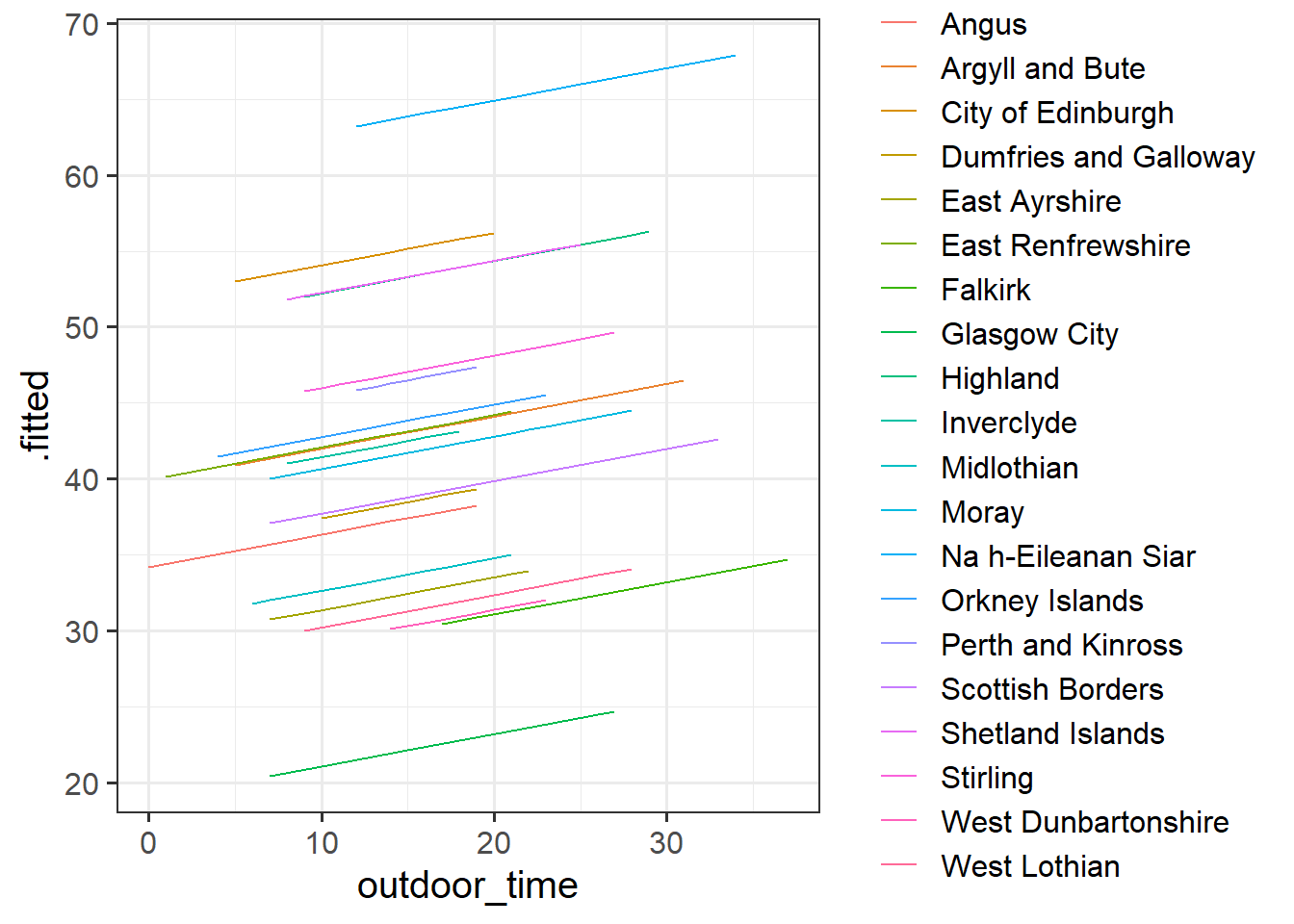
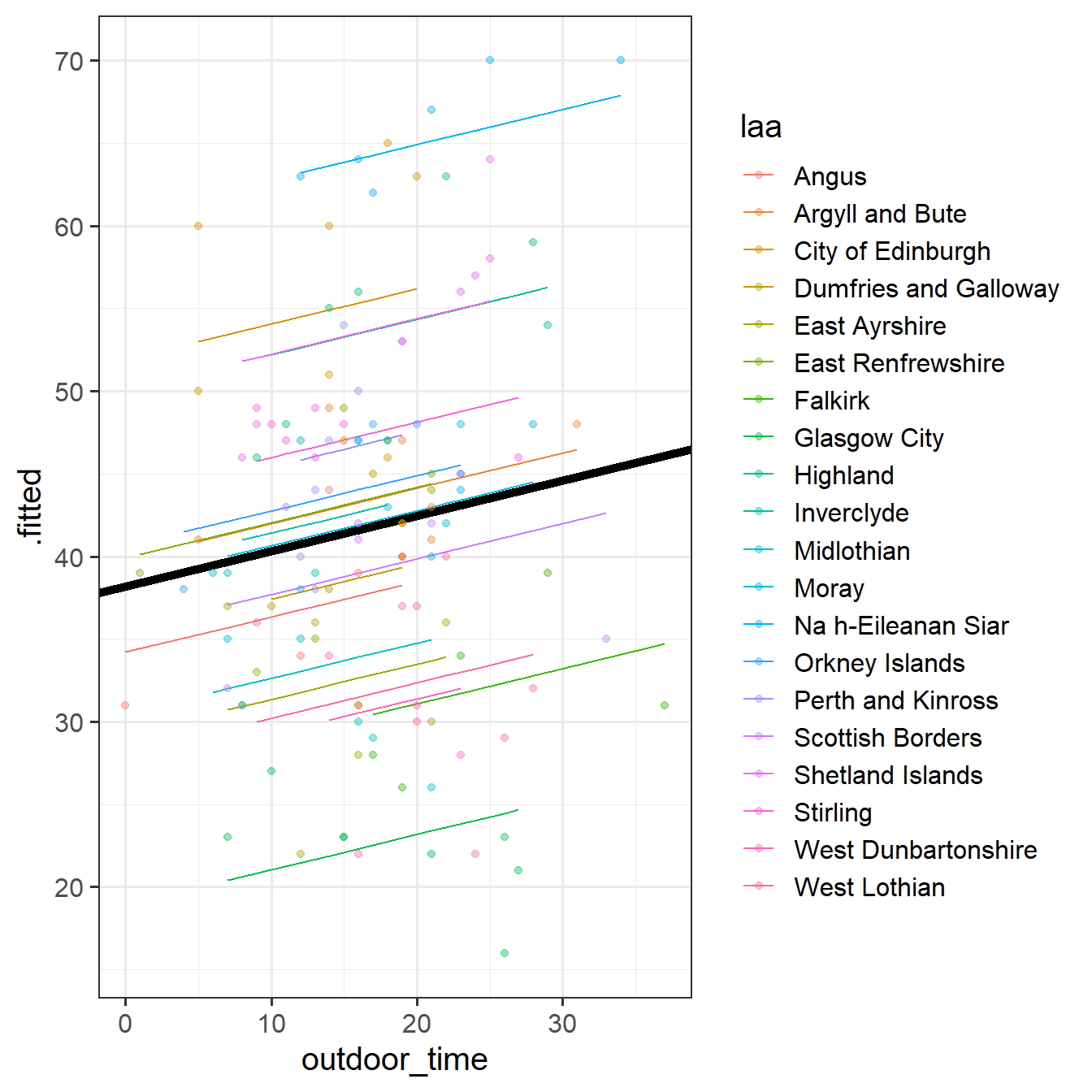
# .weights <dbl>, .wtres <dbl>Add to the code below to plot the model fitted values, and color them according to LAA. (you will need to edit ri_model to be whatever name you assigned to your model).
augment(ri_model) %>%
ggplot(aes(x = outdoor_time, y = ......
We have just fitted the model: \[ \begin{align} & \text{For person } j \text{ in LAA } i \\ & \color{red}{\textrm{Wellbeing}_{ij}}\color{black} = \color{blue}{\beta_{0i} \cdot 1 + \beta_{1} \cdot \textrm{Outdoor Time}_{ij}}\color{black} + \varepsilon_{ij} \\ & \color{blue}{\beta_{0i}}\color{black} = \gamma_{00} + \color{orange}{\zeta_{0i}} \\ \end{align} \]
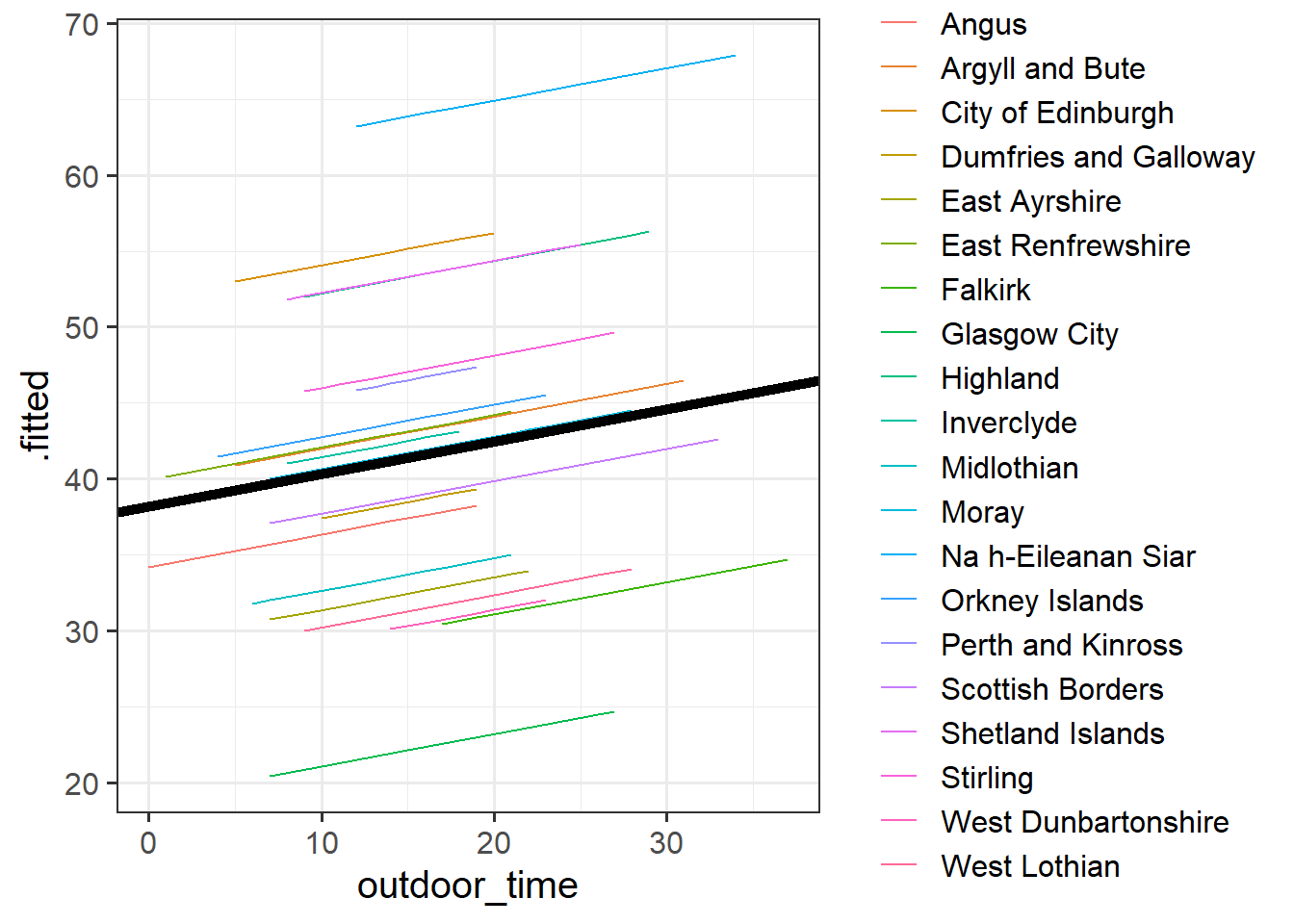
For our estimates of \(\gamma_{00}\) (the fixed value around which LAA intercepts vary) and \(\beta_1\) (the fixed estimate of the relationship between wellbeing and outdoor time), we can use fixef().
fixef(ri_model) (Intercept) outdoor_time
38.189795 0.213492 Can you add to the plot in the previous question, a thick black line with the intercept and slope given by fixef()?
Hint: geom_abline()
lmer(wellbeing~1 + outdoor_time + (1|laa),
data = scotmw)
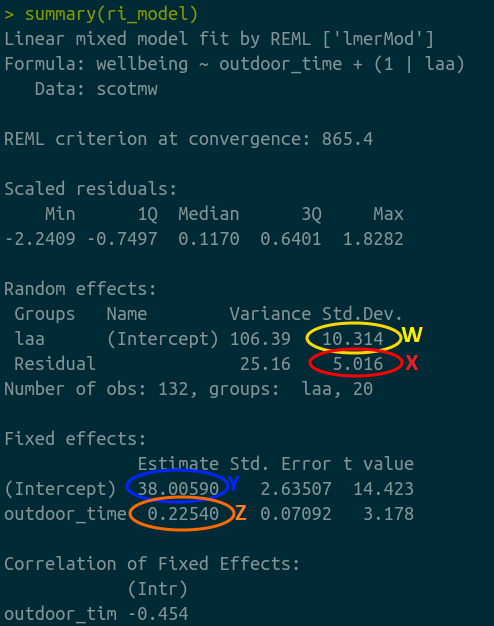
Match the coloured sections Yellow (W), Red (X), Blue (Y), and Orange (Z) in Figure 5 to the descriptions below of Figure 4 A through D.
- where the black line cuts the y axis
- the standard deviation of the distances from all the individual LAA lines to the black line
- the slope of the black line
- the standard deviation of the distances from all the individual observations to the line for the LAA to which it belongs.
Match the colours sections and descriptions from the previous question, to the mathematical terms in the model equation:
\[ \begin{align} & \text{Level 1:} \\ & \color{red}{Wellbeing_{ij}}\color{black} = \color{blue}{\beta_{0i} \cdot 1 + \beta_{1} \cdot OutdoorTime_{ij}}\color{black} + \varepsilon_{ij} \\ & \text{Level 2:} \\ & \color{blue}{\beta_{0i}}\color{black} = \gamma_{00} + \color{orange}{\zeta_{0i}} \\ \quad \\ & \text{where} \\ & \color{orange}{\zeta_0}\color{black} \sim N(0, \sigma_{\color{orange}{\zeta_{0}}}\color{black}) \text{ independently} \\ & \varepsilon \sim N(0, \sigma_{\varepsilon}) \text{ independently} \\ \end{align} \]
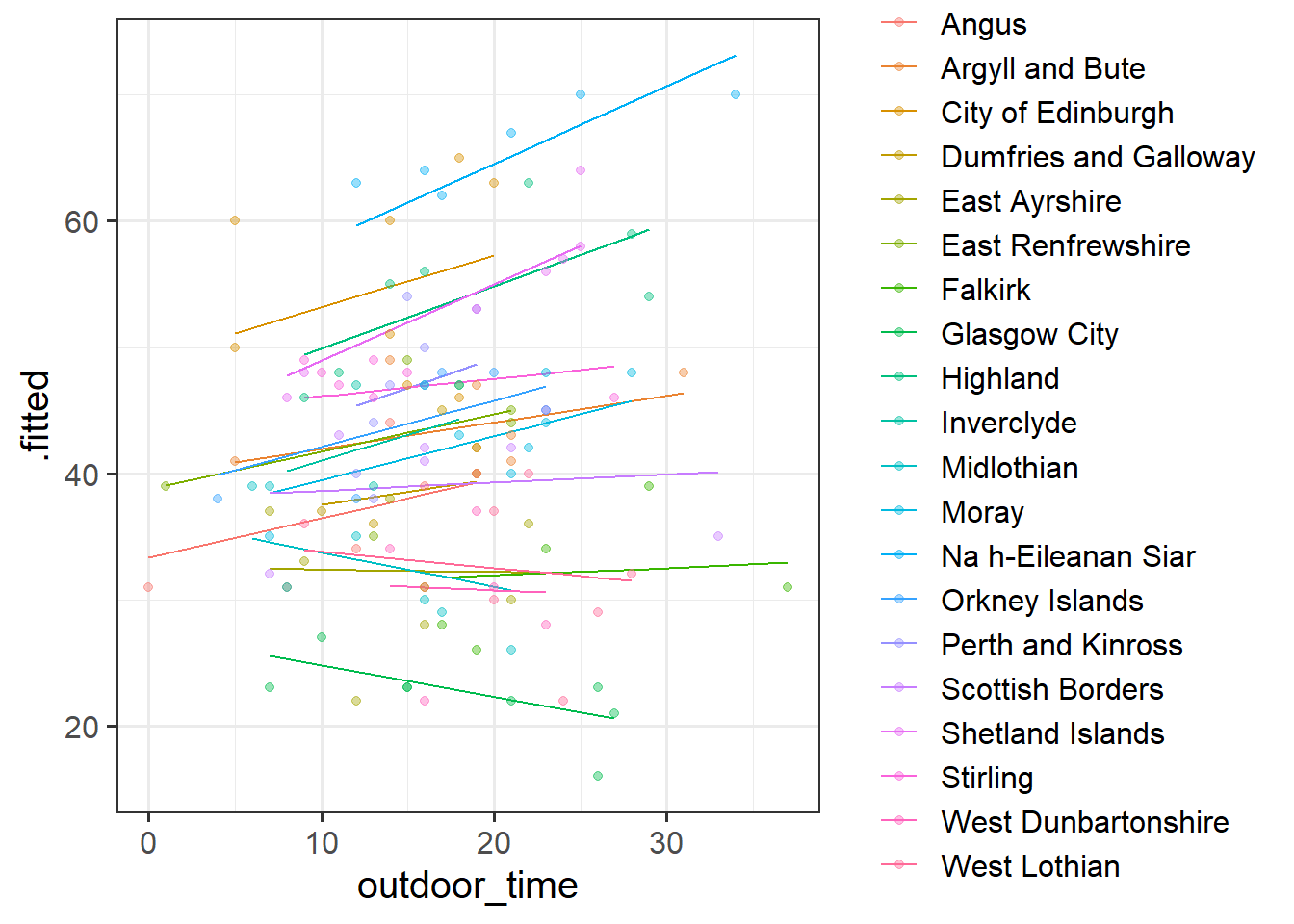
Fit a model which allows also (along with the intercept) the effect of outdoor_time to vary by-LAA.
Then, using augment() again, plot the model fitted values. What do you think you will see?
Does it look like this model better represents the individual LAAs? Take a look at, for instance, Glasgow City.
Exercises: Repeated Measures
While the wellbeing example considers the groupings or ‘clusters’ of different LAAs, a more common grouping in psychological research is that of several observations belonging to the same individual. One obvious benefit of this is that we can collect many more observations with fewer participants but control for the resulting dependency of observations.
Data: REPLICATION Audio/SDMT study
Recall the data from the previous week, from an experiment in which executive functioning was measured (via the SDMT) for people when listening to different types of audio, either via normal speakers or via noise-cancelling headphones.
This week, we have data from a replication of that study, in which the researchers managed to recruit 30 participants. Unfortunately, some participants did not complete all the trials, so we have an unbalanced design. The data is available at https://uoepsy.github.io/data/ef_replication.csv.
| variable | description |
|---|---|
| PID | Participant ID |
| trial_n | Trial Number (1-15) |
| audio | Audio heard during the test ('no_audio', 'white_noise','music') |
| headphones | Whether the participant listened via speakers in the room or via noise cancelling headphones |
| SDMT | Symbol Digit Modalities Test (SDMT) score |
How many participants are there in the data?
How many have complete data (15 trials)?
What is the average number of trials that participants completed? What is the minimum?
Does every participant have some data for each type of audio?
The count() function will likely be useful here.
The model below is sometimes referred to as the “null model” (or “intercept only model”). The grouping structure of the data is specified in the model, but nothing more.
nullmod <- lmer(SDMT ~ 1 + (1 | PID), data = efrep)Fit the model and examine the summary.
How much of the variation in SDMT scores is down to participant grouping?
Set the reference levels of the audio and headphones variables to “no audio” and “speakers” respectively.
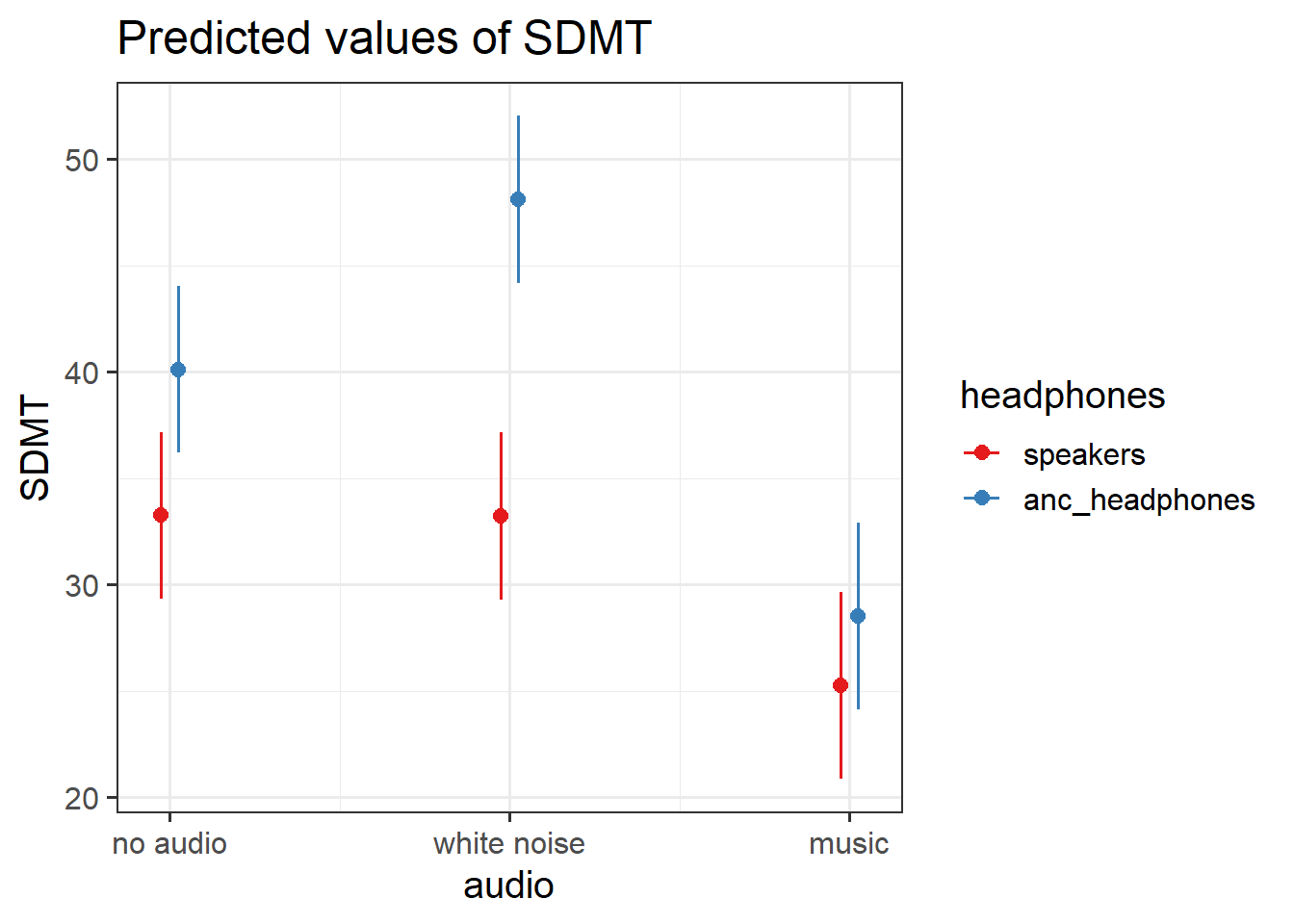
Fit a multilevel model to address the research question below.
How do different types of audio interfere with executive functioning, and does this interference differ depending upon whether or not noise-cancelling headphones are used?
- what is our outcome variable of interest?
- what are our predictor variables (and interactions?) that we are interested in?
- these should be in the fixed effects part.
- these should be in the fixed effects part.
- what is the clustering?
- this should be the random effects
(1 | cluster)part
- this should be the random effects
- does audio type (
audio) vary within clusters, or between?- if so, we might be able to fit a random slope of
audio | cluster. if not, then it doesn’t make sense to do so.
- if so, we might be able to fit a random slope of
- does delivery mode (
headphones) vary within clusters, or between? - if so, we might be able to fit a random slope ofheadphones | cluster. if not, then it doesn’t make sense to do so.
If you get an error about model convergence, consider changing the optimiser (see the “model estimation” box)
We now have a model, but we don’t have any p-values, confidence intervals, or inferential criteria on which to draw conclusions.
Pick a method of your choosing and perform a test of/provide an interval for the relevant effect of interest.
Provide a brief write-up of the results along with a visualisation.
As with normal regression, we have two main ways in which we can conduct inference. We can focus on our coefficients, or we can compare models.2
There are a whole load of different methods available for drawing inferences from multilevel models, which means it can be a bit of a never-ending rabbit hole. For the purposes of this course, we’ll limit ourselves to these two:
| df approximations | likelihood-based | |
|---|---|---|
| tests or CIs for model parameters | library(parameters)model_parameters(model, ci_method="kr") |
confint(model, type="profile") |
| model comparison (different fixed effects, same random effects) |
library(pbkrtest)KRmodcomp(model1,model0) |
anova(model0,model) |
fit models with REML=TRUE.good option for small samples |
fit models with REML=FALSE.needs large N at both levels (40+) |
Exercises: Longitudinal
Another very crucial advantage of these methods is that we can use them to study how people change over time.
Data: Wellbeing in Work
The “Wellbeing in Work” dataset contains information on employees who were randomly assigned to one of three employment conditions:
- control: No change to employment. Employees continue at 5 days a week, with standard allocated annual leave quota.
- unlimited_leave : Employees were given no limit to their annual leave, but were still expected to meet required targets as specified in their job description.
- fourday_week: Employees worked a 4 day week for no decrease in pay, and were still expected to meet required targets as specified in their job description.
Wellbeing was was assessed at baseline (start of maintenance), 12 months post, 24 months post, and 36 months post.
The researchers had two main questions:
- Q1): Overall, did the participants’ wellbeing stay the same or did it change?
- Q2): Did the employment condition groups differ in the how wellbeing changed over the assessment period?
The data is available, in .rda format, at https://uoepsy.github.io/data/wellbeingwork3.rda. You can read it directly into your R environment using:
load(url("https://uoepsy.github.io/data/wellbeingwork3.rda"))After running the code above you will find the data in an object called wellbeingwork3 in your environment.
Q1): Overall, did the participants’ wellbeing stay the same or did it change?
Each of our participants have measurements at 4 assessments. We need to think about what this means for the random effects that we will include in our model (our random effect structure). Would we like our models to accommodate individuals to vary in their overall wellbeing, to vary in how they change in wellbeing over the course of the assessment period, or both?
To investigate whether wellbeing changed over the course of the assessments, or whether it stayed the same, we can fit and compare 2 models:
- A null model.
- A model with wellbeing predicted by time point.
And we can then compare them in terms of model fit (as mentioned above, there are lots of different ways we might do this).
Our sample size here (180 participants, each with 4 observations) is reasonably large given the relative simplicity of our model. We might consider running a straightforward Likelihood Ratio Test using anova(restricted_model, full_model) to compare our two models (in which case we should fit them with REML=FALSE)
- Remember, we shouldn’t use likelihood ratio tests to compare models with different random effect structures.
- (For now, don’t worry too much about “singular fits”. We’ll talk more about how we might deal with them next week!)
Q: Did the employment condition groups differ in the how wellbeing changed over the assessment period?
It helps to break it down. There are two questions here:
- do groups differ overall?
- do groups differ over time?
We can begin to see that we’re asking two questions about the Condition variable here: “is there an effect of Condition?” and “Is there an interaction between TimePoint and Condition?”.
Try fitting two more models which incrementally build these levels of complexity, and compare them (perhaps to one another, perhaps to models from the previous question - think about what each comparison is testing!)
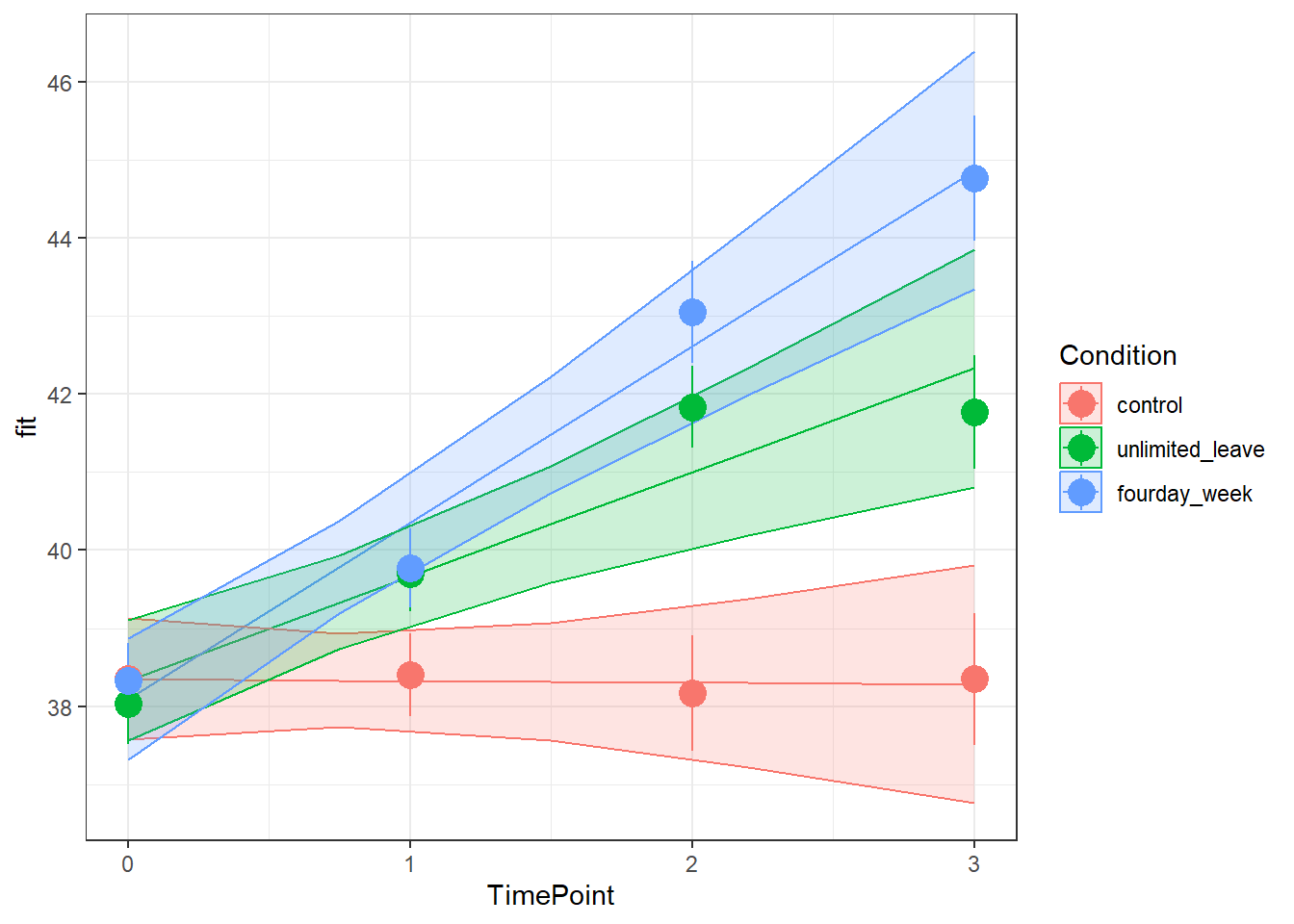
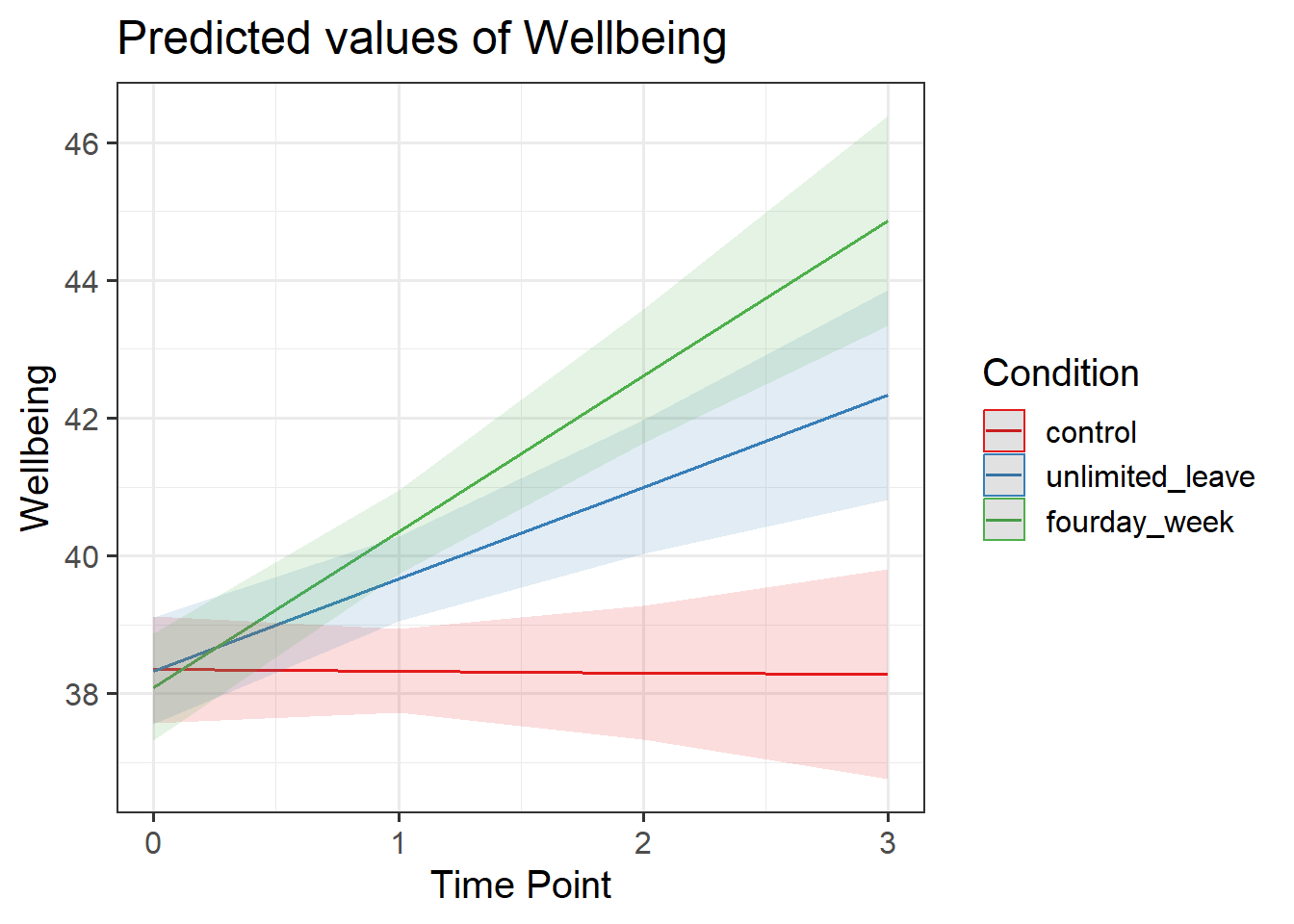
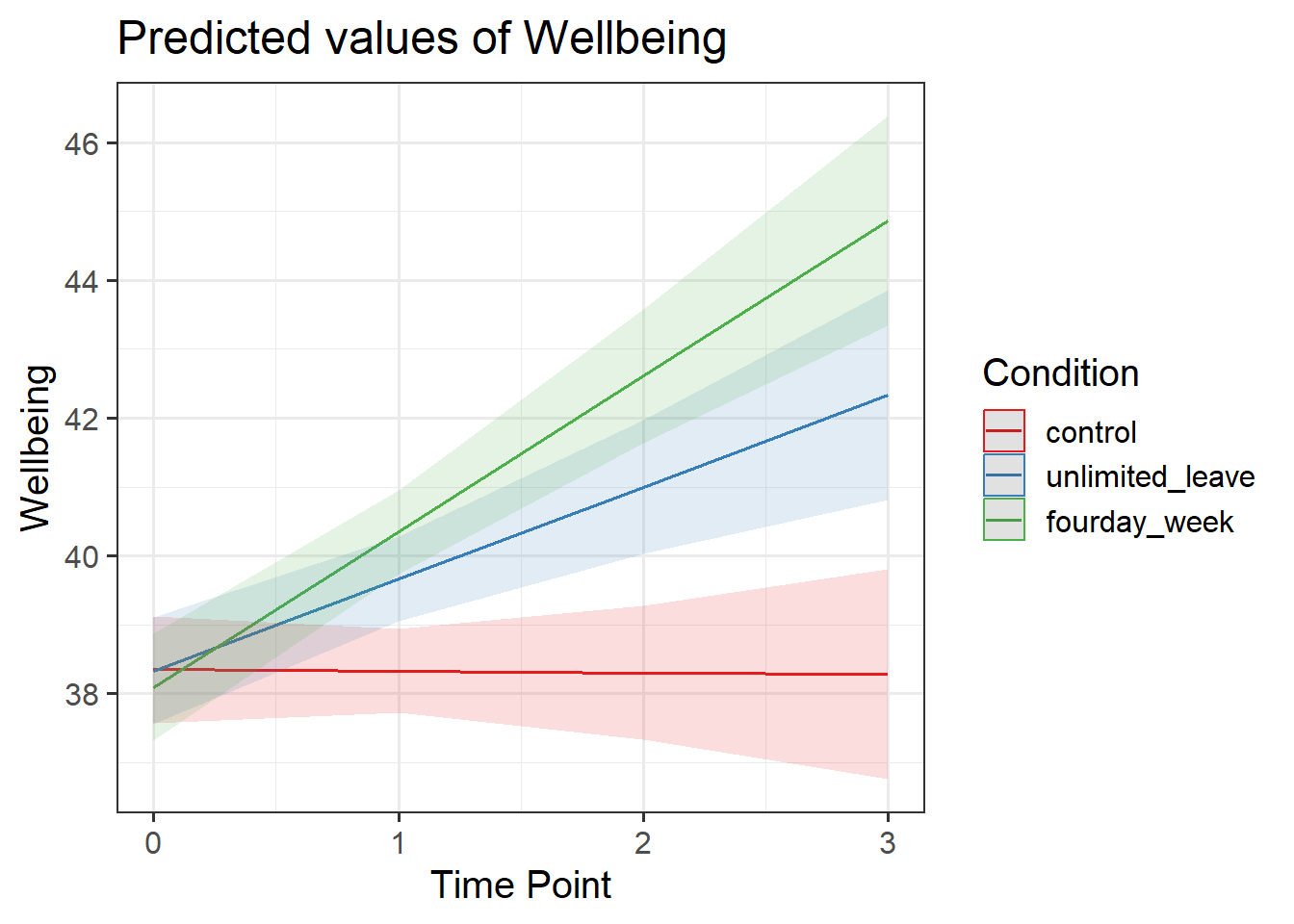
Visualise the model estimated change in wellbeing over time for each Condition.
There are lots of ways you can visualise the model, try a couple:
- Using the effects package, this might help:
as.data.frame(effect("TimePoint*Condition", model))
- We can also use sjPlot, as we have seen in DAPR2
Examine the parameter estimates and interpret them (i.e., what does each parameter represent?
Can you match them with parts of the plot obtained from plot_model(m.full, type="int")?
We can get the fixed effects using fixef(model), and we can also use tidy(model) from the broom.mixed package, and similar to lm models in DAPR2, we can pull out the bit of the summary() using summary(model)$coefficients.
Footnotes
“(g)lmer” here stands for “(generalised) linear mixed effects regression”.↩︎
What is the difference between testing coefficients and comparing models? This is most easily seen when the model includes a categorical variable as a predictor. The
summary()function would return, for each level of the predictor (excluding the reference level), a coefficient, its standard error, the t-statistic, and the p-value for a test of whether the coefficient is significantly different from 0. A model comparison between that model and a null model without the categorical predictor would collapse all levels into a single test across all levels of the predictor.↩︎