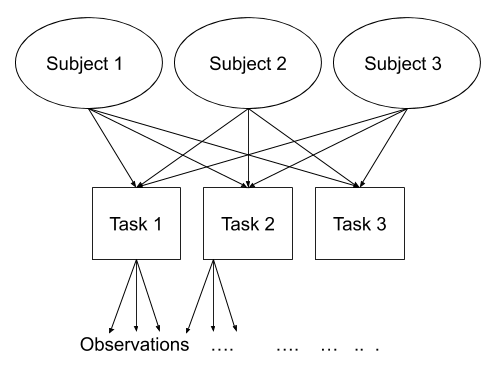
Recap of multilevel models
No exercises this week
This week, there aren’t any exercises, but there is a small recap reading of multilevel models, followed by some ‘flashcard’ type boxes to help you test your understanding of some of the key concepts.
Please use the lab sessions to go over exercises from previous weeks, as we all as asking any questions about the content below.
School Example
Do children scores in maths improve more in school 2020 vs school 2040?
Consider the following data, representing longitudinal measurements on 10 students from an urban public primary school. The outcome of interest is mathematics achievement. The data were collected at the end of first grade and annually thereafter up to sixth grade, but not all students have six observations. The variable year has been mean-centred to have mean 0 so that results will have as baseline the average.
How many students in each school?
## schoolid
## 2020 2040 Sum
## 21 21 42We have 42 students, 21 in school with id 2020 and 21 in school with id 2040:
The number of observations per child are as follows.
table(data$childid)##
## 253404261 253413681 270199271 273026452 273030991 273059461 278058841 285939962
## 6 3 3 3 5 5 5 3
## 288699161 289970511 292017571 292020281 292020361 292025081 292026211 292027291
## 5 5 3 6 6 5 5 5
## 292027531 292028181 292028931 292029071 292029901 292033851 292772811 293550291
## 5 5 5 5 5 5 3 4
## 295341521 298046562 299680041 301853741 303652591 303653561 303654611 303658951
## 6 3 5 3 5 3 5 3
## 303660691 303662391 303663601 303668751 303671891 303672001 303672861 303673321
## 4 6 6 5 5 3 3 4
## 307407931 307694141
## 3 4We can see that for some children we have fewer than the 6 observations: some have 3, 4, or 5.
School 2020
Let’s start by considering only the children in school 2020. The mathematics achievement over time is shown, for each student, in the plot below:
data2020 <- data %>%
filter(schoolid == 2020)
ggplot(data2020, aes(x = year, y = math)) +
geom_point() +
facet_wrap(~ childid, labeller = label_both) +
labs(x = "Year (mean centred)", y = "Maths achievement score")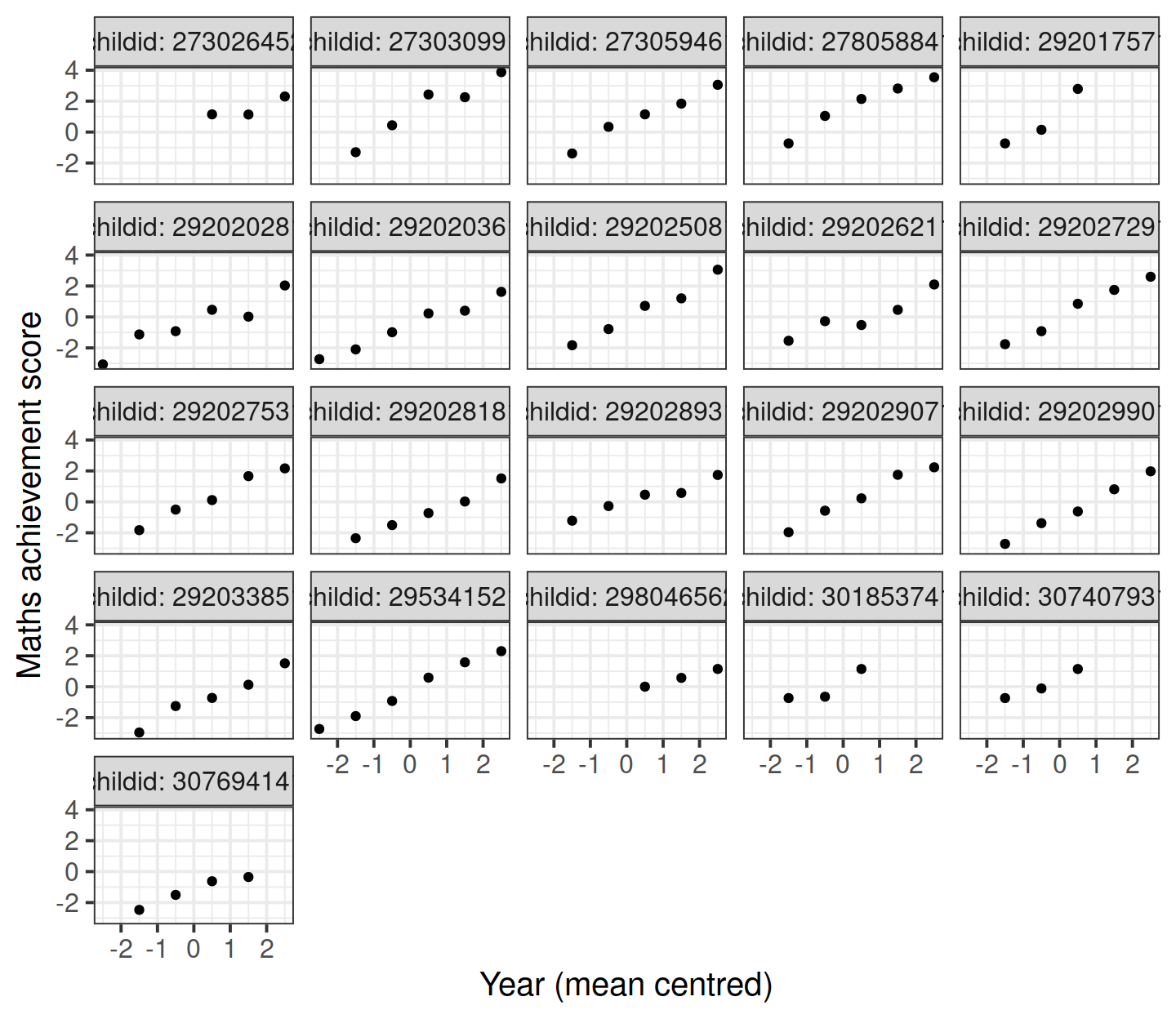
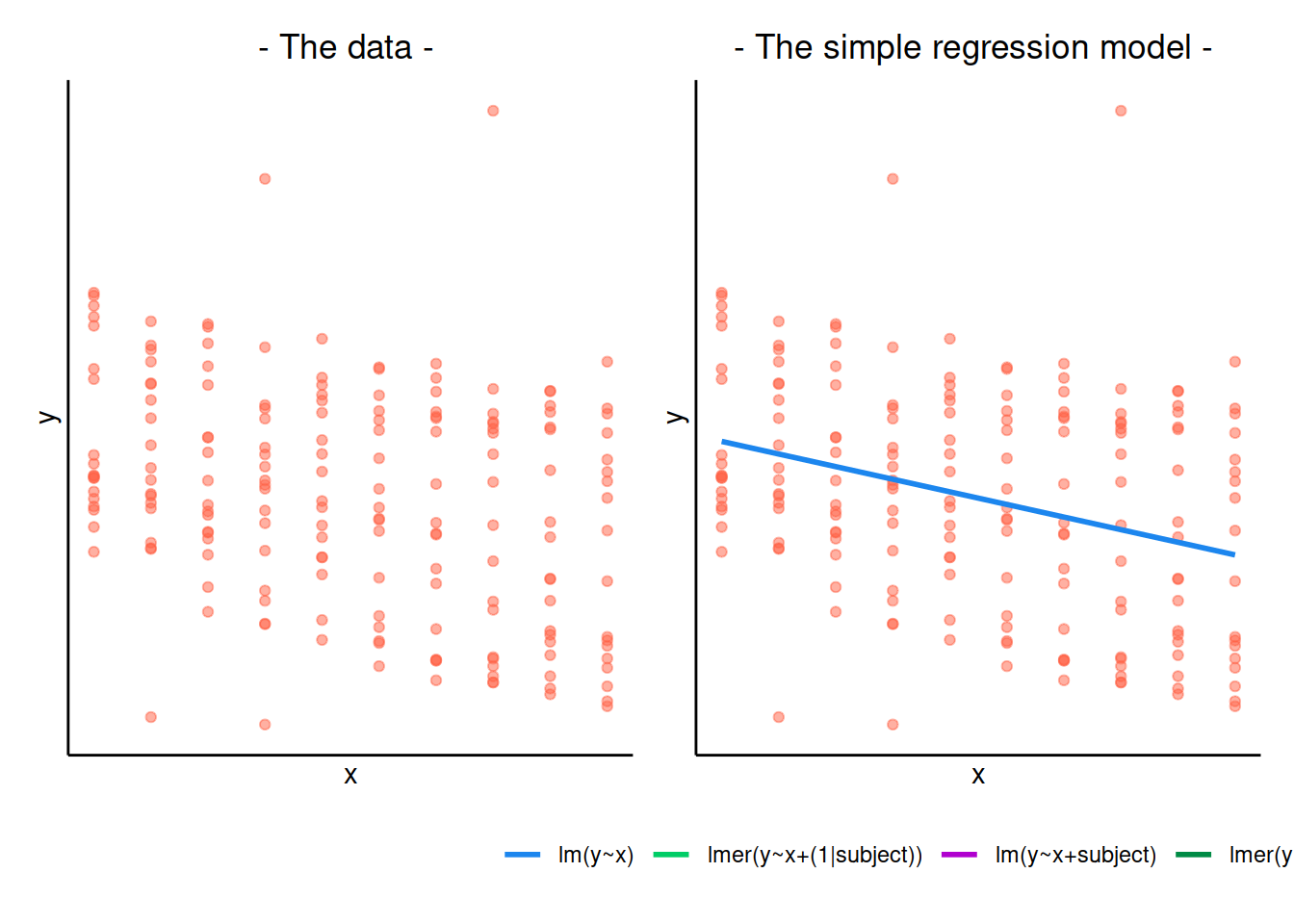
Clearly, the measurements of mathematics achievement related to each student are grouped data as they refer to the same entity.
If we were to ignore this grouping and consider all children as one single population, we would obtain misleading results.The observations for the same student are clearly correlated. Some students consistently have a much better performance than other students, perhaps due to underlying numerical skills.
A fundamental assumption of linear regression models is that the residuals, and hence the data too, should be uncorrelated. In this example this is not the case.
The following plot considers all data as a single population
ggplot(data2020, aes(x = year, y = math)) +
geom_point() +
geom_smooth(method = lm, se = FALSE) +
labs(x = "Year (mean centred)", y = "Maths achievement score")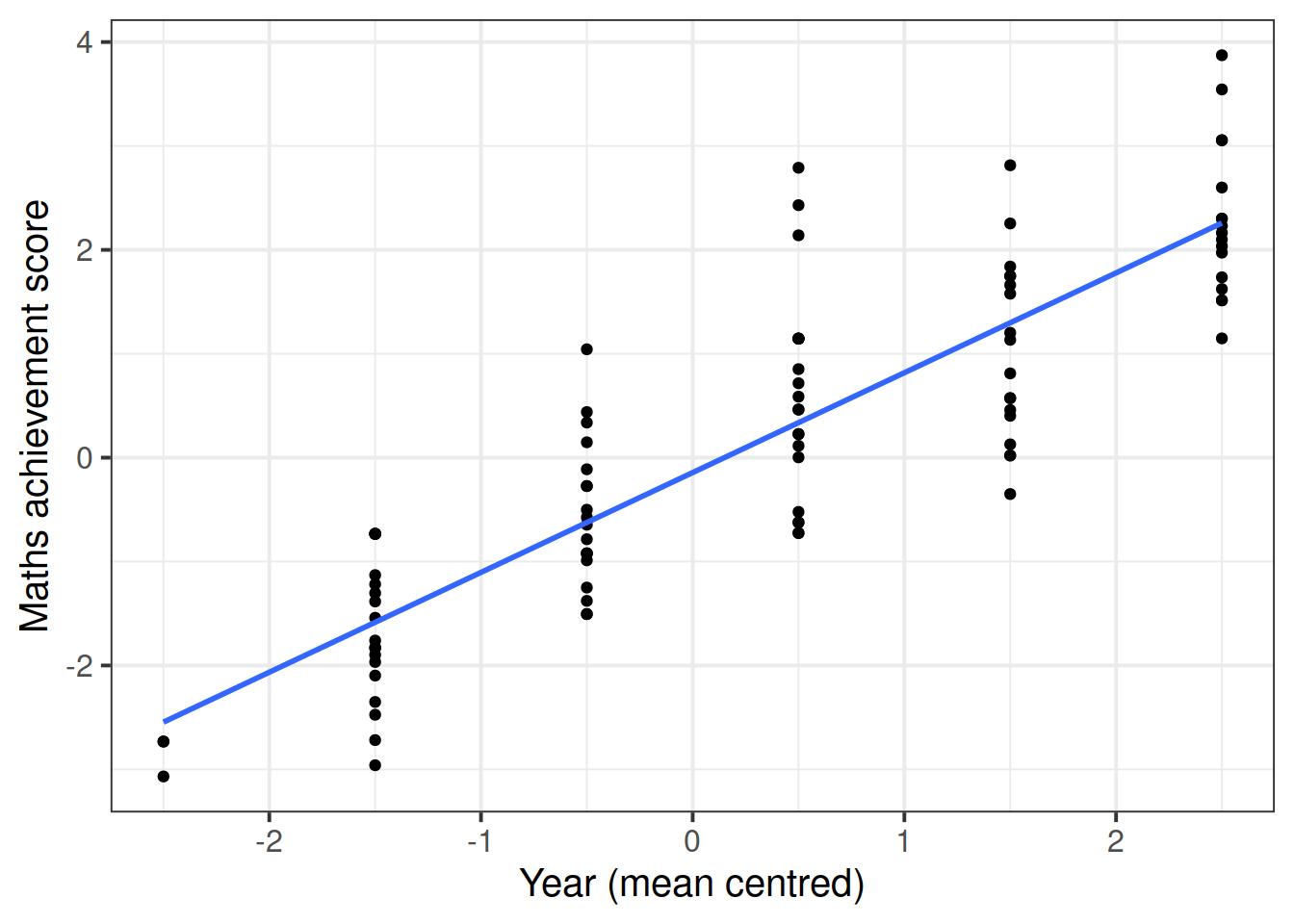
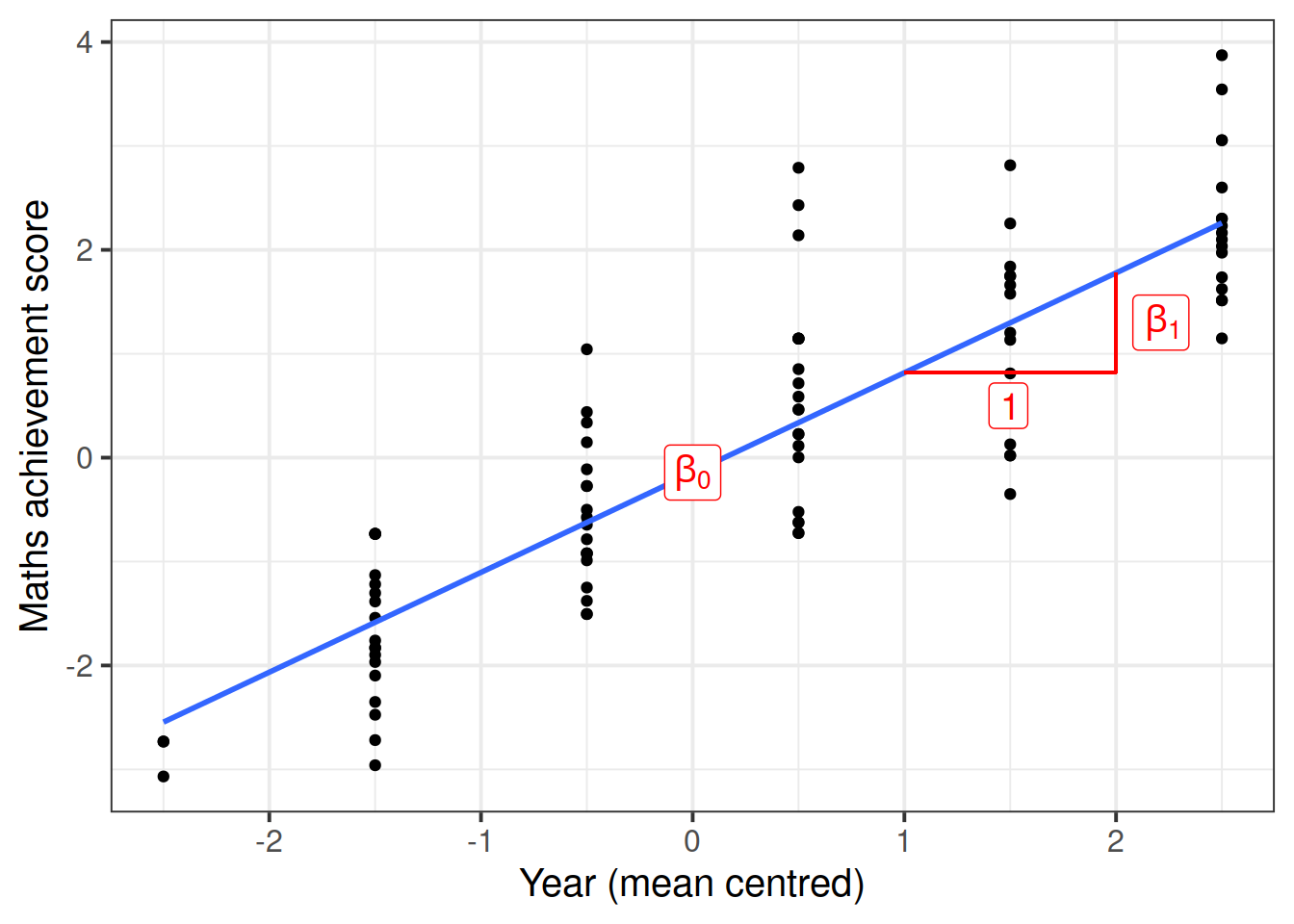
This is a simple linear regression model for the mathematics measurement of individual \(i\) on occasion \(j\): \[ \text{math}_{ij} = \beta_0 + \beta_1 \ \text{year}_{ij} + \epsilon_{ij} \]
where the subscript \(ij\) denotes the \(j\)th measurement from child \(i\).
Let’s fit this in R
m0 <- lm(math ~ year, data = data2020)
summary(m0)##
## Call:
## lm(formula = math ~ year, data = data2020)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.6478 -0.6264 -0.1101 0.4543 2.4529
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.14323 0.08493 -1.687 0.095 .
## year 0.96072 0.05662 16.968 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.8126 on 95 degrees of freedom
## Multiple R-squared: 0.7519, Adjusted R-squared: 0.7493
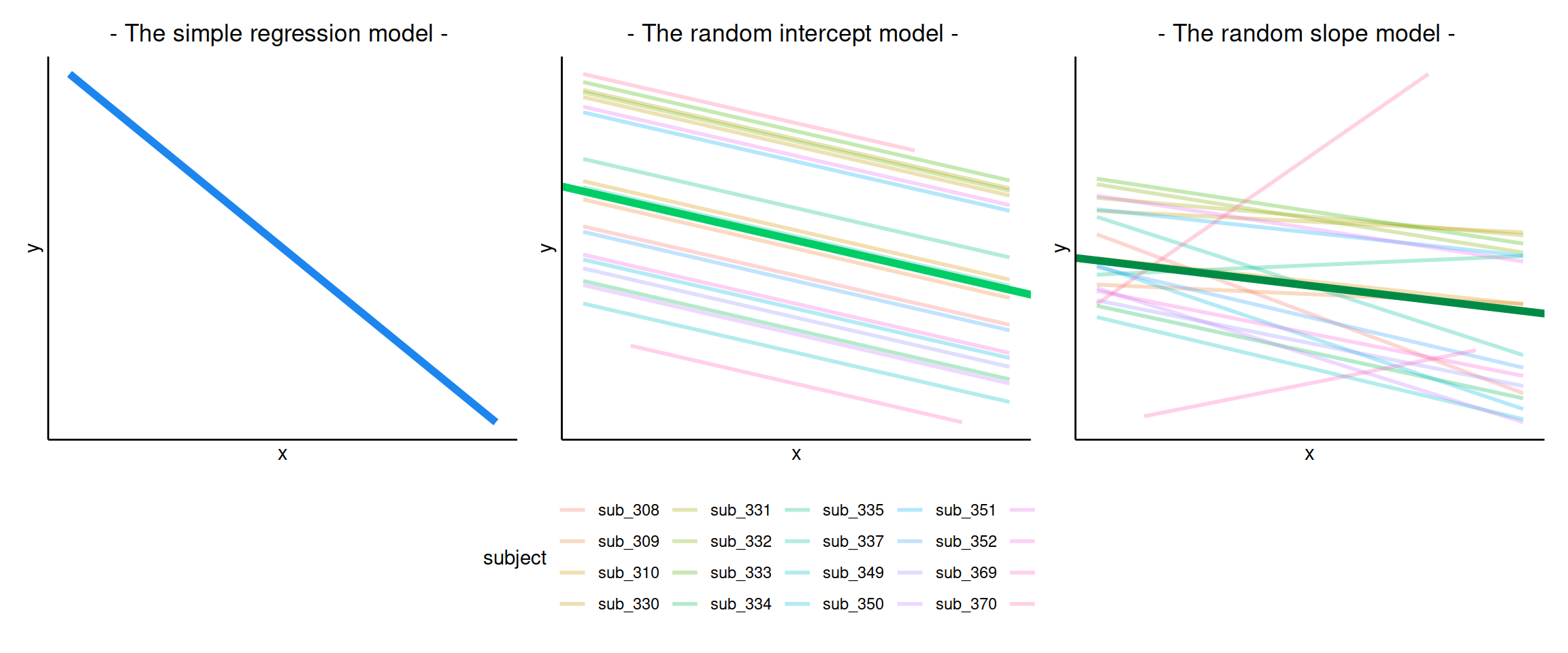
## F-statistic: 287.9 on 1 and 95 DF, p-value: < 2.2e-16The intercept and slope of this model can be visually represented as:
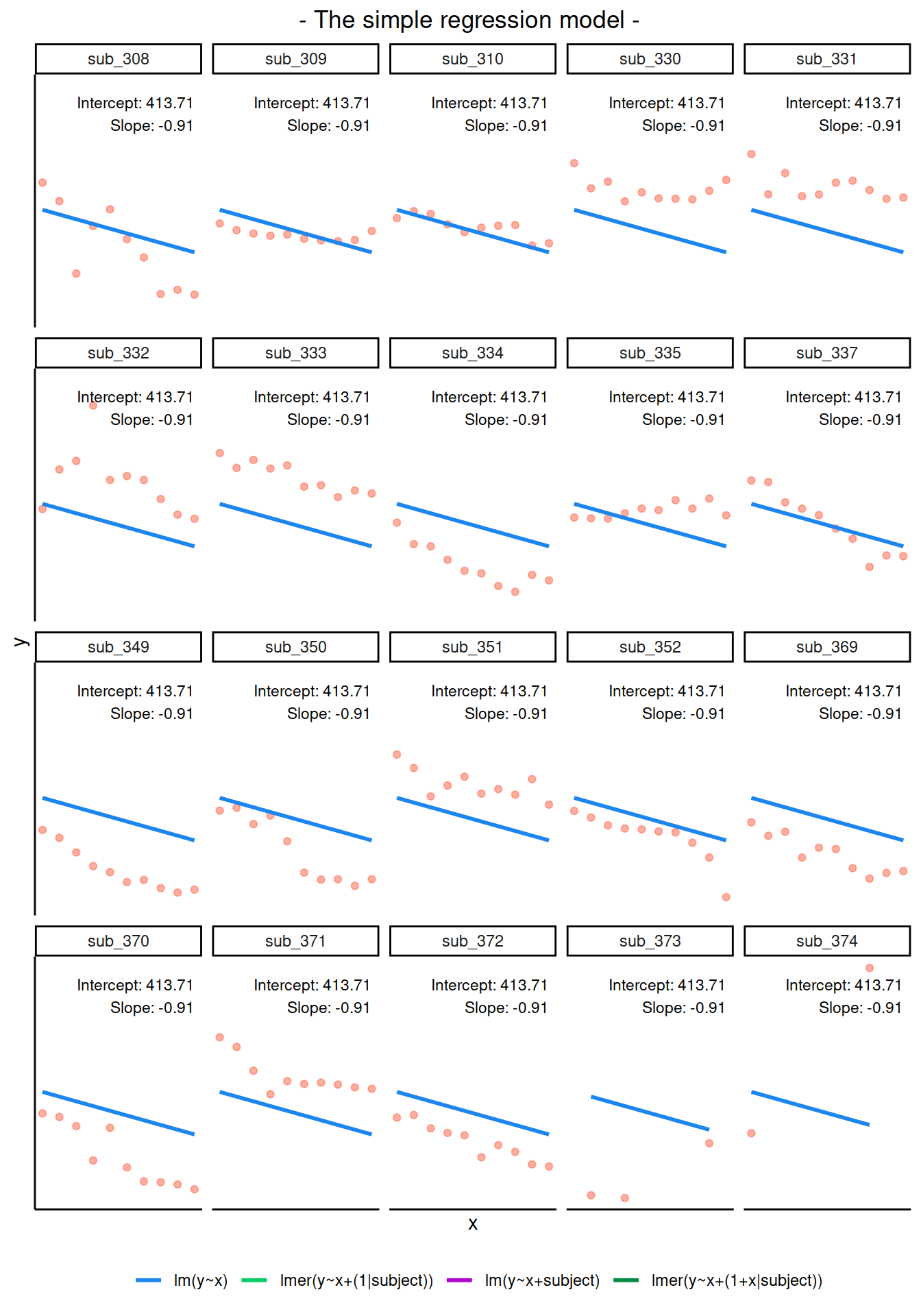
Random intercept and slopes
In reality, we see that each student has their own line, with a different intercept and slope. In other words, they all have different values of maths achievement when year = 0 and they also differ in their learning rate.
ggplot(data2020, aes(x = year, y = math, color = childid)) +
geom_point() +
geom_smooth(method = lm, se = FALSE, fullrange = TRUE,
size = 0.5) +
labs(x = "Year (mean centred)", y = "Maths achievement score") +
theme(legend.position = 'bottom')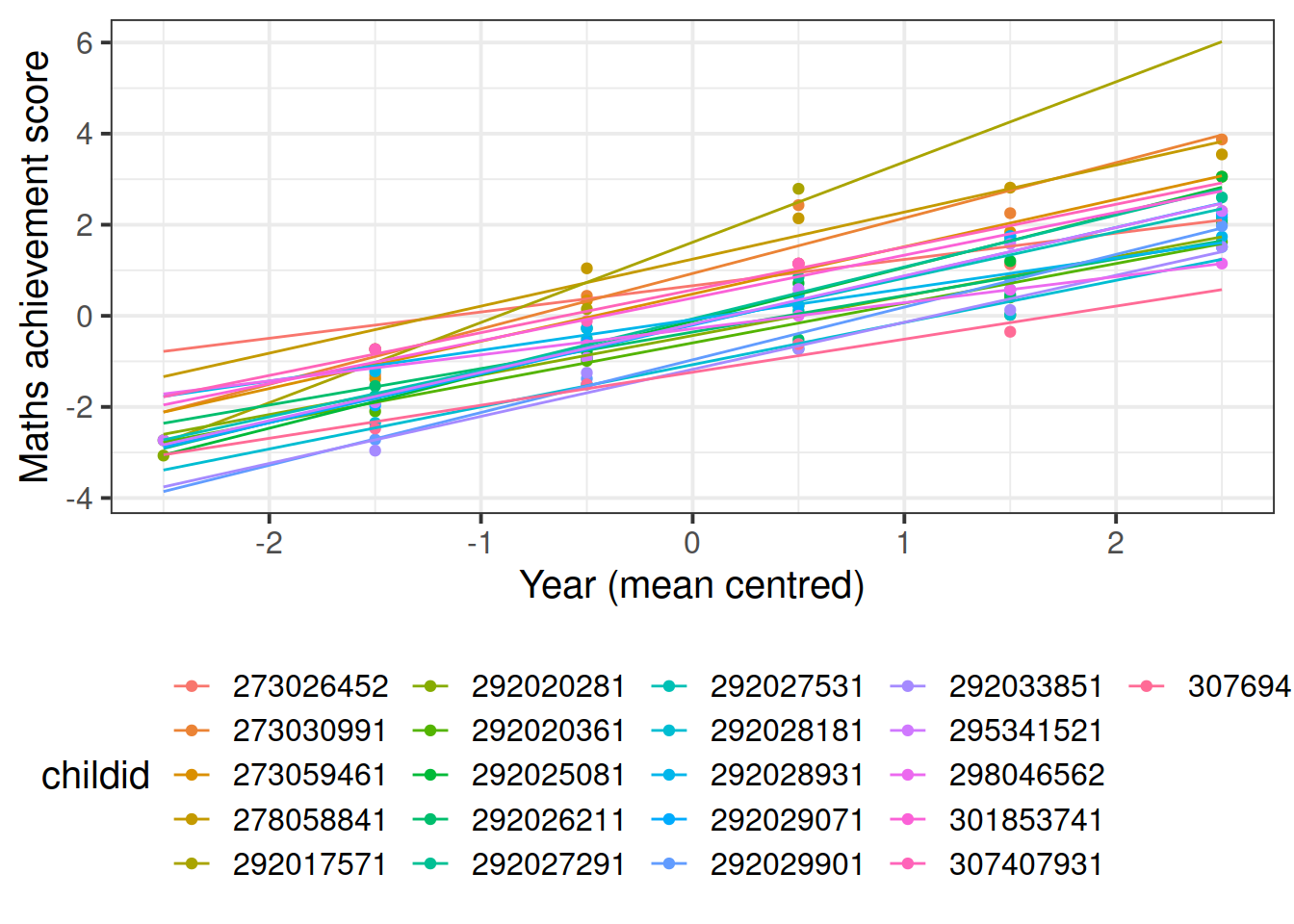
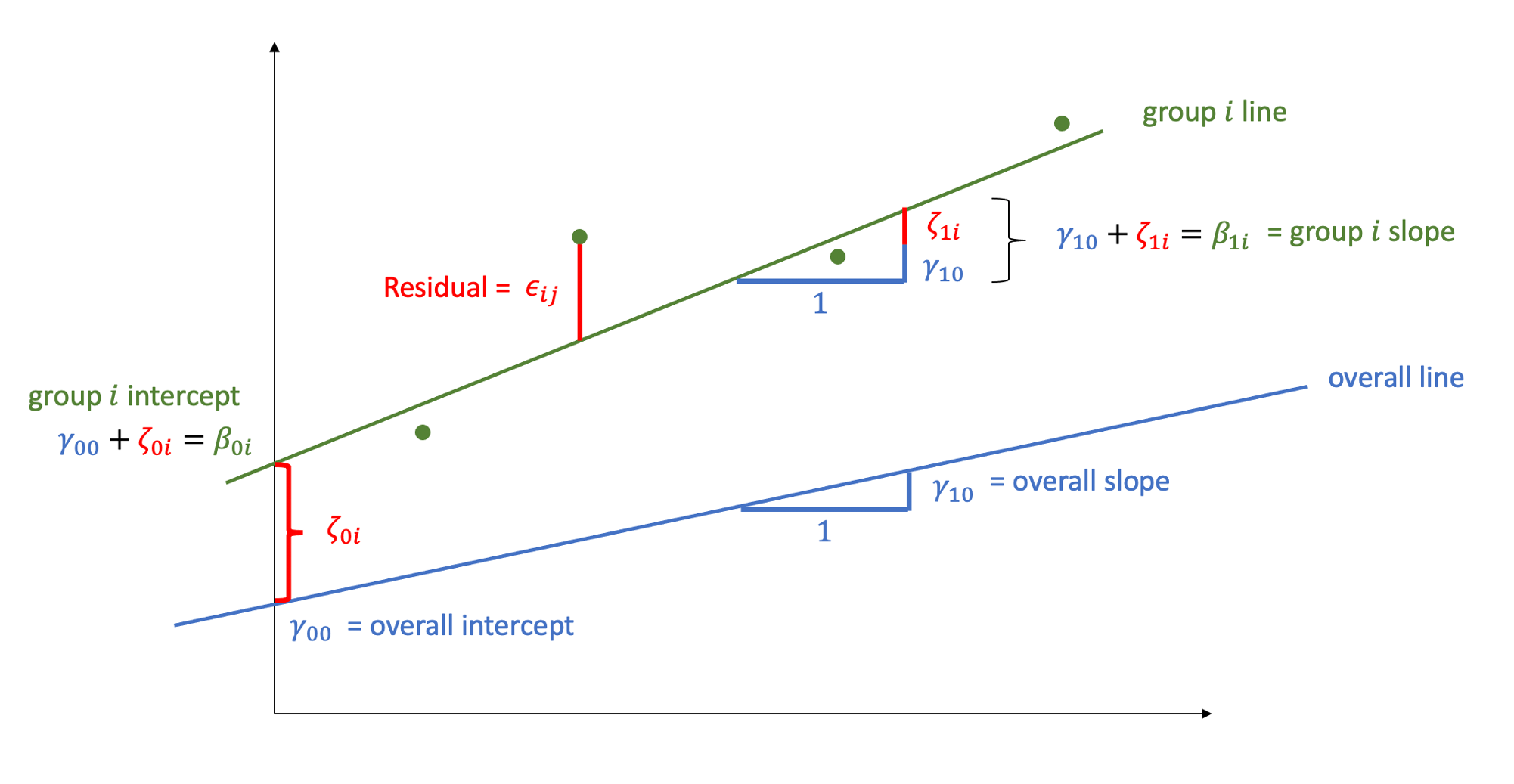
Let’s now write a model where each student has their own intercept and slope: \[ \begin{aligned} \text{math}_{ij} &= \beta_{0i} + \beta_{1i} \ \text{year}_{ij} + \epsilon_{ij} \\ &= (\text{intercept for child } i) + (\text{slope for child } i) \ \text{year}_{ij} + \epsilon_{ij} \\ &= (\gamma_{00} + \zeta_{0i}) + (\gamma_{10} + \zeta_{1i}) \ \text{year}_{ij} + \epsilon_{ij} \end{aligned} \]
where
\(\beta_{0i}\) is the intercept of the line for child \(i\)
\(\beta_{1i}\) is the slope of the line for child \(i\)
\(\epsilon_{ij}\) are the deviations of each child’s measurement \(\text{math}_{ij}\) from the line of child \(i\)
We can think each child-specific intercept (respectively, slope) as being made up of two components: an “overall” intercept \(\gamma_{00}\) (slope \(\gamma_{10}\)) and a child-specific deviation from the overall intercept \(\zeta_{0i}\) (slope \(\zeta_{1i}\)):
\(\beta_{0i} = \gamma_{00} + \zeta_{0i} = \text{(overall intercept) + (deviation for child }i)\)
\(\beta_{1i} = \gamma_{10} + \zeta_{1i} = \text{(overall slope) + (deviation for child }i)\)
FACT
Deviations from the mean average to zero (and sum to zero too!)
As you know, deviations from the mean average to 0.
This holds for the errors \(\epsilon_{ij}\), as well as the deviations \(\zeta_{0i}\) from the overall intercept, and the deviations \(\zeta_{1i}\) from the overall slope.
Think of data \(y_1, ..., y_n\) and their mean \(\bar y\). The average of the deviations from the mean is \[ \begin{aligned} \frac{\sum_i (y_i - \bar y)}{n} = \frac{\sum_i y_i }{n} - \frac{\sum_i \bar y}{n} = \bar y - \frac{n * \bar y}{n} = \bar y - \bar y = 0 \end{aligned} \]
The child-specific deviations \(\zeta_{0i}\) from the overall intercept are normally distributed with mean \(0\) and variance \(\sigma_0^2\). Similarly, the deviations \(\zeta_{1i}\) of the slope for child \(i\) from the overall slope come from a normal distribution with mean \(0\) and variance \(\sigma_1^2\). The correlation between random intercepts and slopes is \(\rho = \text{Cor}(\zeta_{0i}, \zeta_{1i}) = \frac{\sigma_{01}}{\sigma_0 \sigma_1}\):
\[ \begin{bmatrix} \zeta_{0i} \\ \zeta_{1i} \end{bmatrix} \sim N \left( \begin{bmatrix} 0 \\ 0 \end{bmatrix}, \begin{bmatrix} \sigma_0^2 & \rho \sigma_0 \sigma_1 \\ \rho \sigma_0 \sigma_1 & \sigma_1^2 \end{bmatrix} \right) \]
The random errors, independently from the random effects, are distributed \[ \epsilon_{ij} \sim N(0, \sigma_\epsilon^2) \]
This is fitted using lmer():
library(lme4)
m1 <- lmer(math ~ 1 + year + (1 + year | childid), data = data2020)
summary(m1)## Linear mixed model fit by REML ['lmerMod']
## Formula: math ~ 1 + year + (1 + year | childid)
## Data: data2020
##
## REML criterion at convergence: 166.6
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.3119 -0.6125 -0.0726 0.6002 2.4197
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## childid (Intercept) 0.50065 0.7076
## year 0.01131 0.1063 0.82
## Residual 0.16345 0.4043
## Number of obs: 97, groups: childid, 21
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) -0.1091 0.1605 -0.68
## year 0.9940 0.0381 26.09
##
## Correlation of Fixed Effects:
## (Intr)
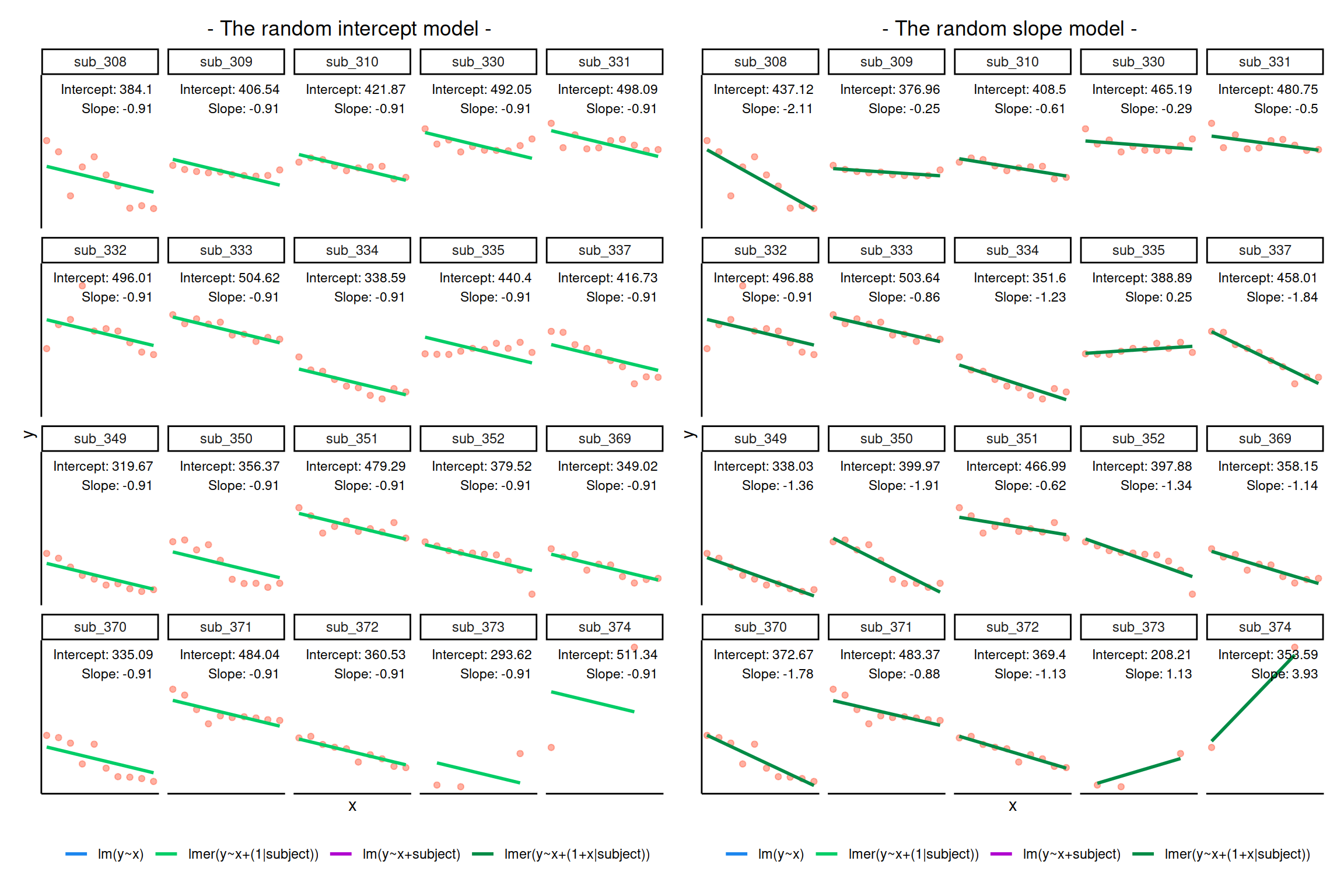
## year 0.433The summary of the lmer output returns estimated values for
Fixed effects:
- \(\widehat \gamma_{00} = -0.109\)
- \(\widehat \gamma_{10} = 0.994\)
Variability of random effects:
- \(\widehat \sigma_{0} = 0.708\)
- \(\widehat \sigma_{1} = 0.106\)
Correlation of random effects:
- \(\widehat \rho = 0.816\)
Residuals:
- \(\widehat \sigma_\epsilon = 0.404\)
Remember: - The child-specific deviations \(\zeta_{0i}\) from the overall intercept are normally distributed with mean \(0\) and variance \(\sigma_0^2\). - Similarly, the deviations \(\zeta_{1i}\) of the slope for child \(i\) from the overall slope come from a normal distribution with mean \(0\) and variance \(\sigma_1^2\).
We can check that our random effects look :
par(mfrow = c(1,2))
qqnorm(ranef(m1)$childid[, 1], main = "Random intercept")
qqline(ranef(m1)$childid[, 1])
qqnorm(ranef(m1)$childid[, 2], main = "Random slope")
qqline(ranef(m1)$childid[, 2])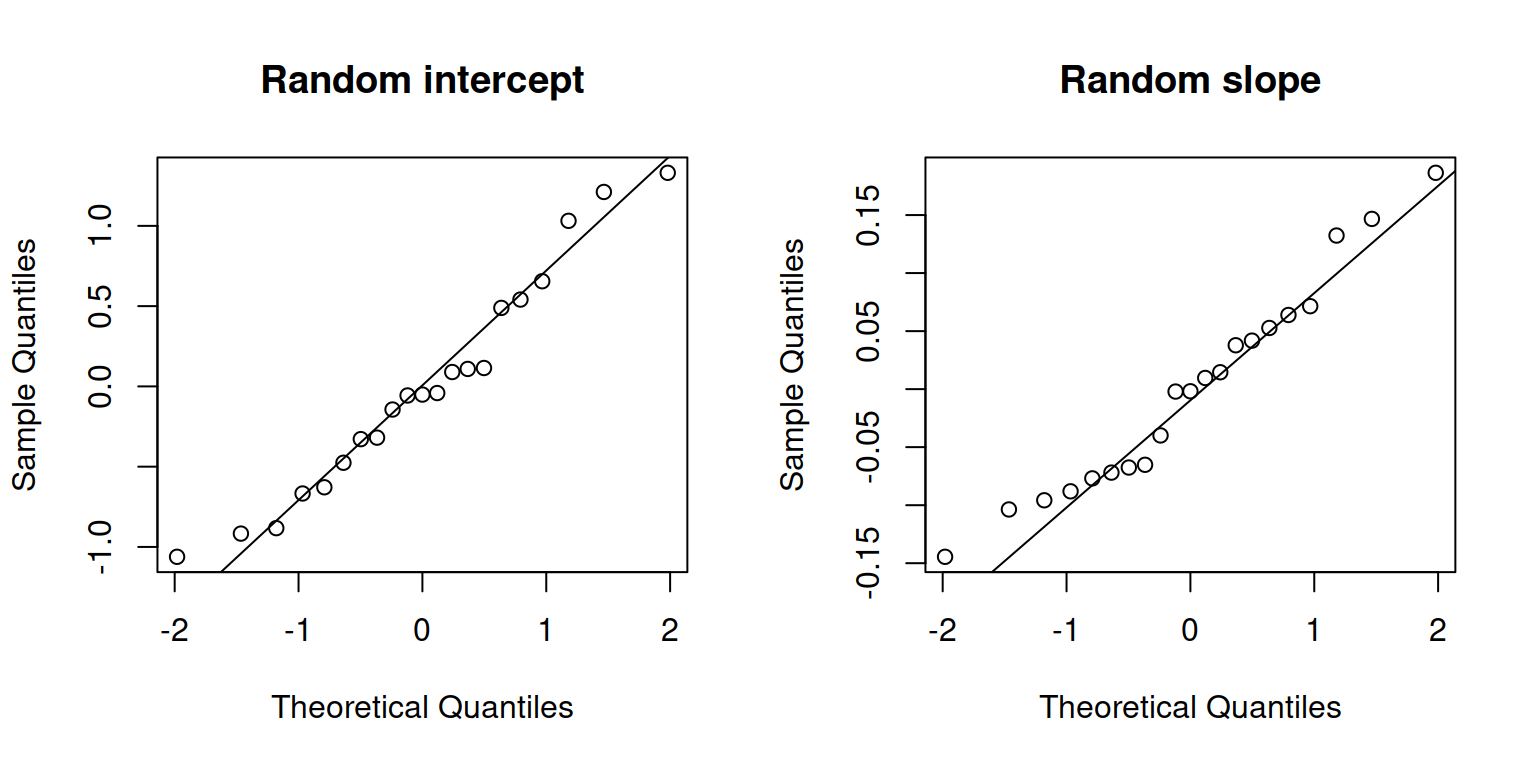
Check normality and independence of errors:
qqnorm(resid(m1), id=0.05)
qqline(resid(m1), id=0.05)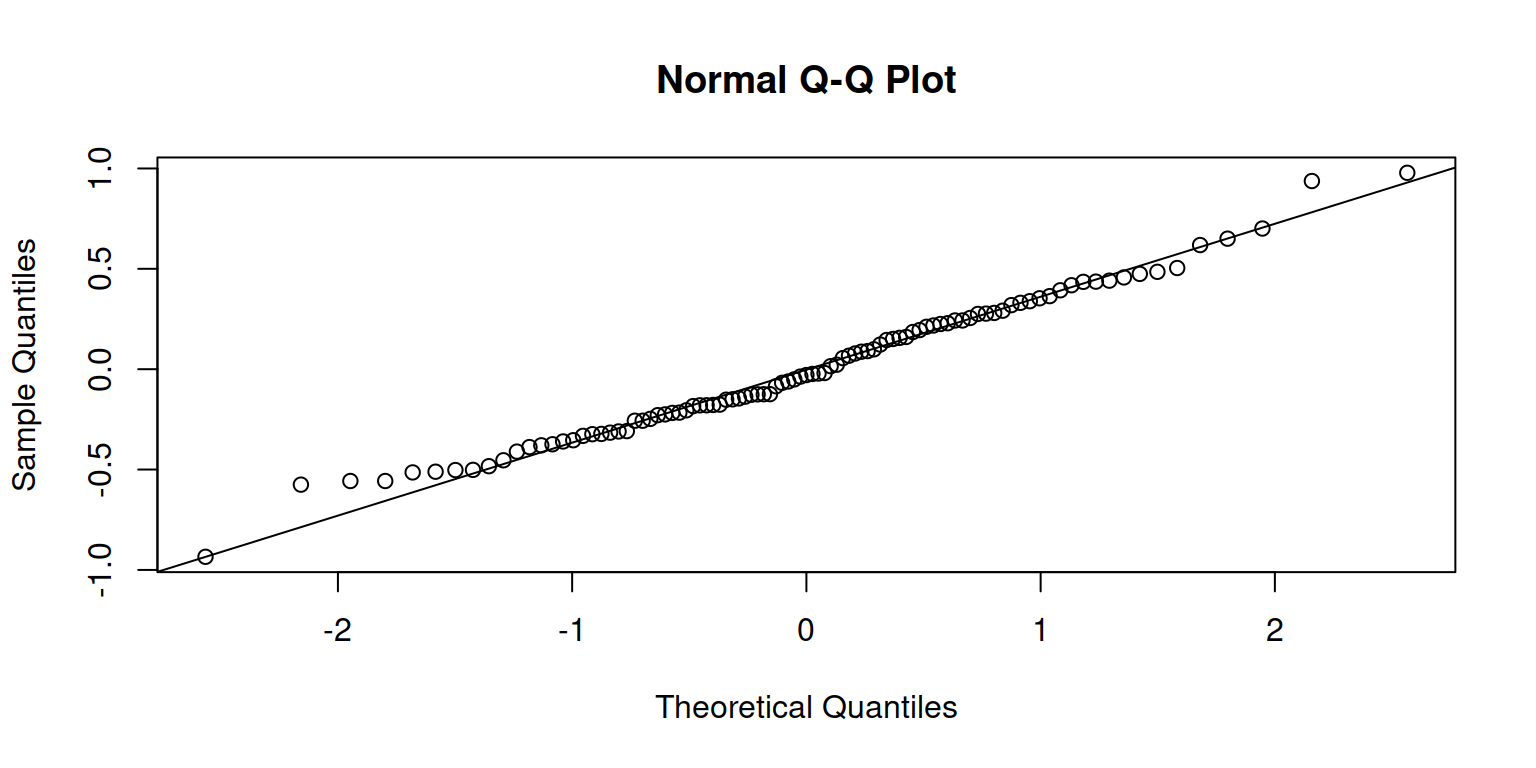
plot(m1,
form = sqrt(abs(resid(.))) ~ fitted(.),
type = c("p","smooth"))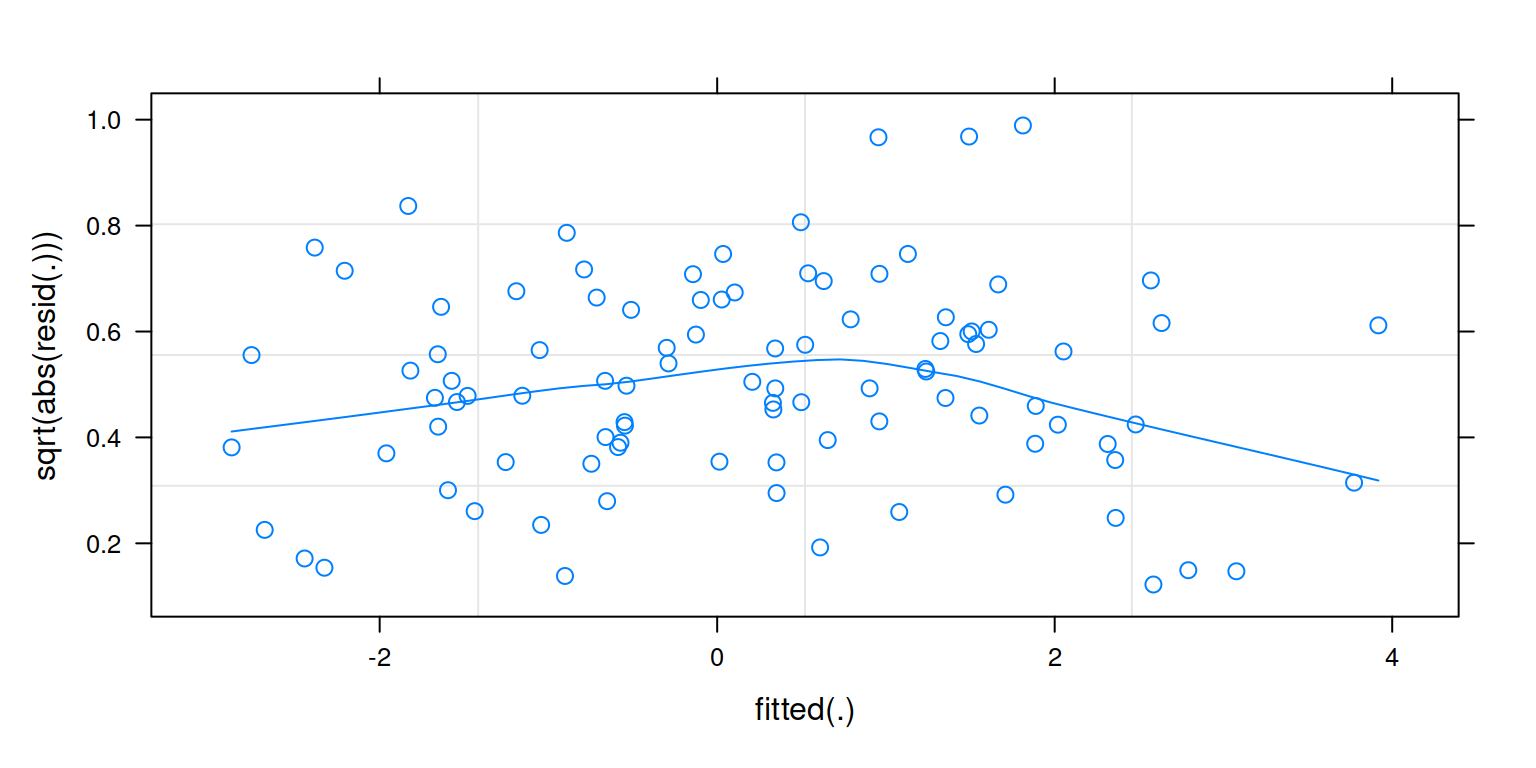
Visually inspect the correlation between the random intercept and slopes:
ggplot(ranef(m1)$childid,
aes(x = `(Intercept)`, y = year)) +
geom_smooth(method = lm, se = FALSE,
color = 'gray', size = 0.5) +
geom_point()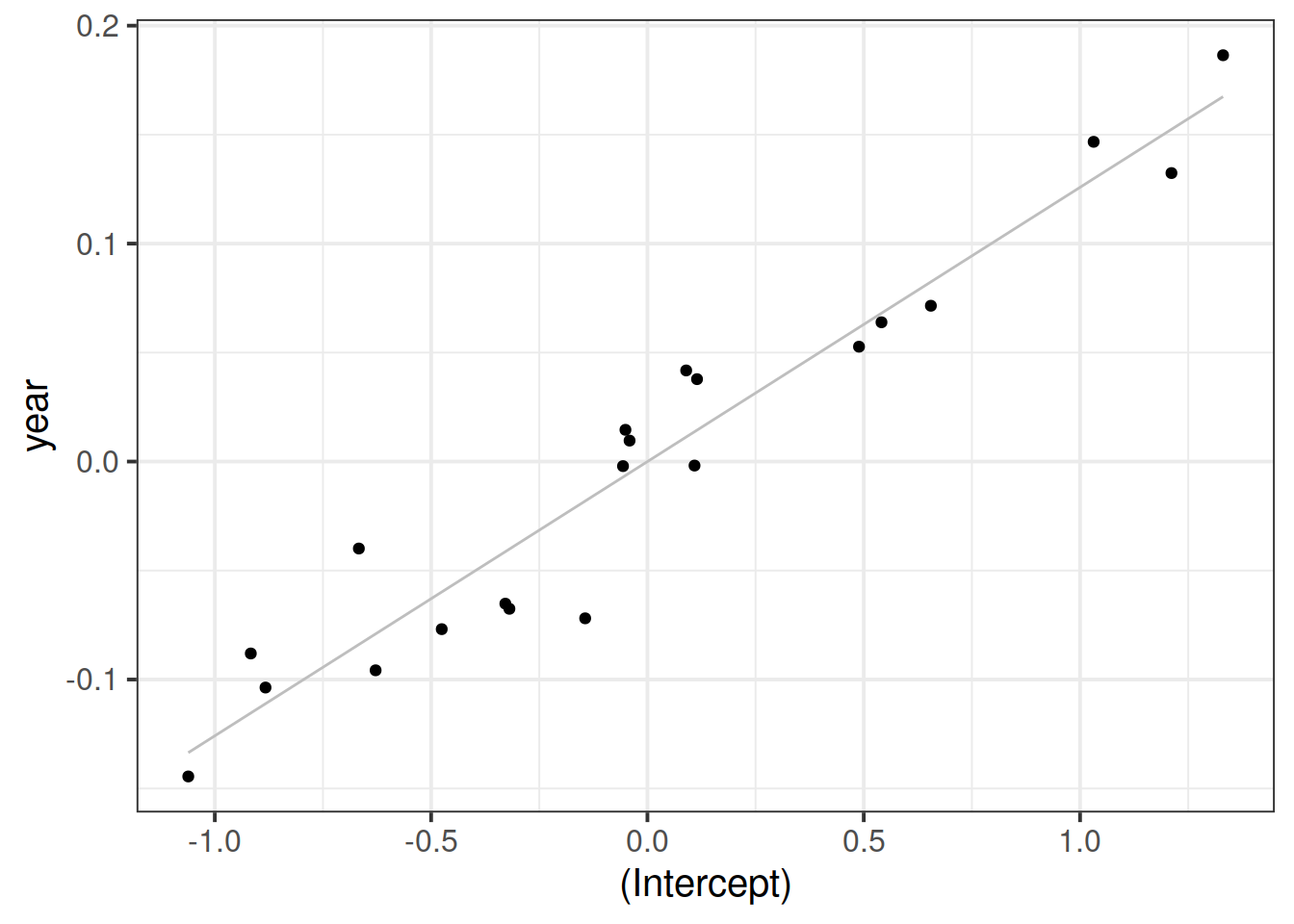
Flashcards: lm to lmer
In a simple linear regression, there is only considered to be one source of random variability: any variability left unexplained by a set of predictors (which are modelled as fixed estimates) is captured in the model residuals.
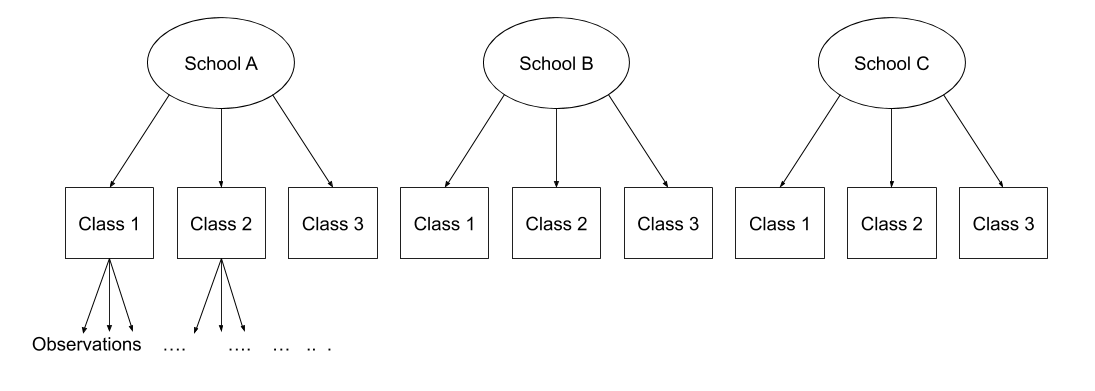
Multi-level (or ‘mixed-effects’) approaches involve modelling more than one source of random variability - as well as variance resulting from taking a random sample of observations, we can identify random variability across different groups of observations. For example, if we are studying a patient population in a hospital, we would expect there to be variability across the our sample of patients, but also across the doctors who treat them.
We can account for this variability by allowing the outcome to be lower/higher for each group (a random intercept) and by allowing the estimated effect of a predictor vary across groups (random slopes).
Before you expand each of the boxes below, think about how comfortable you feel with each concept.
This content is very cumulative, which means often going back to try to isolate the place which we need to focus efforts in learning.