class: center, middle, inverse, title-slide # <b>Week 00: Tidyverse and Markdown Review</b> ## Multivariate Statistics and Methodology using R (MSMR)<br><br> ### Dan Mirman ### Department of Psychology<br>The University of Edinburgh ### AY 2020-2021 --- # Data Visualization Important in *science*, business, journalism, etc. Two key roles: 1. Communication 2. Analysis Summary statistics never give the full *picture*: 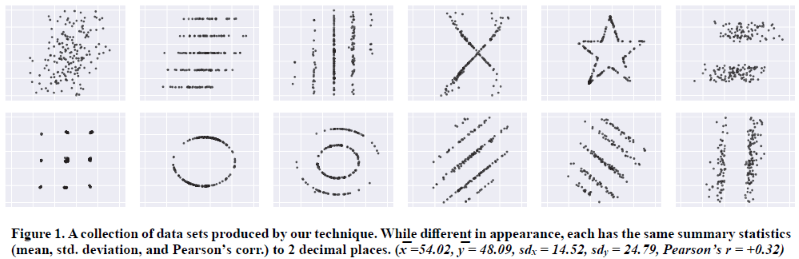 --- # `ggplot`: Grammar of Graphics Many graphing programs treat data visualization like painting picture: red circle at `\(x_1,y_1\)`, blue square at `\(x_2,y_2\)`, etc. This is inefficient and very easy to make mistakes. --- # `ggplot`: Grammar of Graphics Many graphing programs treat data visualization like painting picture: red circle at `\(x_1,y_1\)`, blue square at `\(x_2,y_2\)`, etc. This is inefficient and very easy to make mistakes. ### The `ggplot` way: a graph has 3 core elements 1. A data set 2. A set of mappings between variables in the data set and properties (*aesthetics*) of the graph 3. Layers of *geoms* to instantiate those mappings --- # A simple example A basic line plot: ```r library(ggplot2) ggplot(Orange, aes(x=age, y=circumference, color=Tree)) + geom_line() ``` 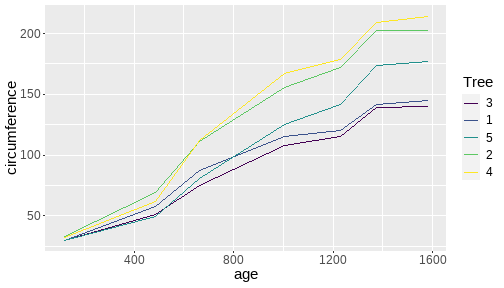<!-- --> --- # A simple example ```r ggplot(Orange, aes(x=age, y=circumference, color=Tree)) + geom_line() + geom_point() # Add points ``` 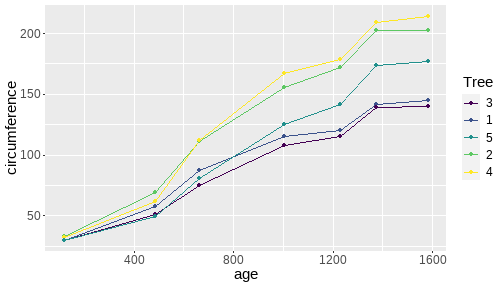<!-- --> Notice that the lines and points have matching colors, but you only need to specify the color mapping once. All geoms will "inherit" the same aesthetic mappings unless otherwise specified. *This helps maintain consistency.* --- # Compute summaries on the fly ```r ggplot(Orange, aes(age, circumference)) + geom_point() + stat_summary(fun=mean, geom="line") ``` 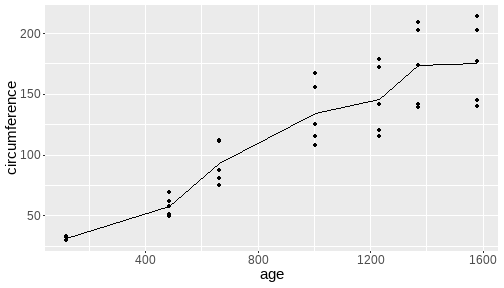<!-- --> Do not need to have different data sets for all data and summarized data. Just use one data set and visualize it in different ways (all observations, grouped by participants, grouped by conditions, etc., etc., ...) *This makes exploratory visualization much easier.* --- # Exclude cases on the fly ```r ggplot(subset(Orange, Tree != "5"), aes(age, circumference)) + geom_point() + stat_summary(fun=mean, geom="line") ``` 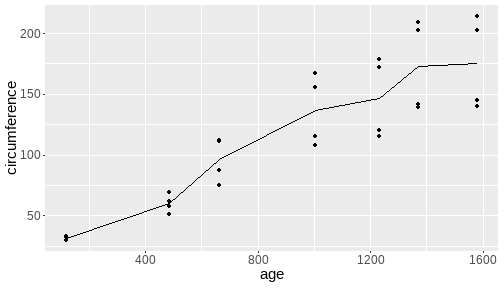<!-- --> This also helps exploratory visualization. --- # Small multiples, aka facets Very important for visualizing complex data sets. The individual facets are properly sized and aligned, and inherit aesthetics to make comparisons easy. ```r ggplot(Orange, aes(age, circumference)) + facet_wrap(~ Tree, nrow=1) + geom_line() ``` 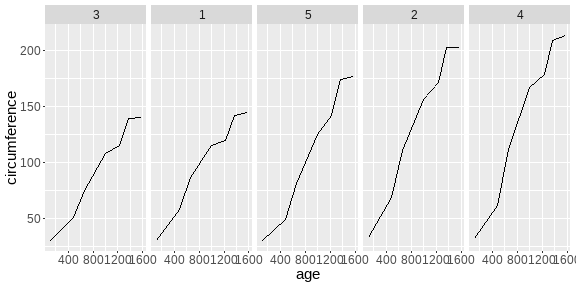<!-- --> --- # Multi-panel plots For combining different plots, use `patchwork` ```r library(patchwork) p1 <- ggplot(esoph, aes(agegp, ncases/ncontrols, color=alcgp)) + stat_smooth(aes(group=alcgp), se=FALSE) + scale_color_brewer(palette = "Set1") p2 <- ggplot(esoph, aes(agegp, ncases/ncontrols, color=tobgp)) + stat_smooth(aes(group=tobgp), se=FALSE) + scale_color_brewer(palette = "Set1") p1 + p2 ``` 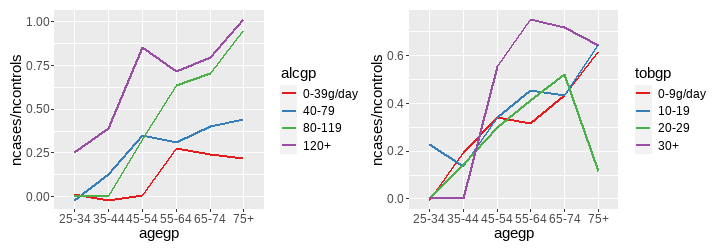<!-- --> --- # Showing variability: Weather data ```r load("./data/Weather_2014.RData") summary(weather_day) ``` ``` ## month day TempAve TempMin ## Min. : 1.000 Min. : 1.00 Min. : 0.6423 Min. :-1.993 ## 1st Qu.: 4.000 1st Qu.: 8.00 1st Qu.: 5.6862 1st Qu.: 3.431 ## Median : 7.000 Median :16.00 Median : 9.0329 Median : 6.486 ## Mean : 6.526 Mean :15.72 Mean : 9.4095 Mean : 6.664 ## 3rd Qu.:10.000 3rd Qu.:23.00 3rd Qu.:13.1557 3rd Qu.:10.230 ## Max. :12.000 Max. :31.00 Max. :19.2297 Max. :14.720 ## TempMax Rain ## Min. : 2.273 Min. : 0.000 ## 1st Qu.: 8.340 1st Qu.: 0.000 ## Median :12.030 Median : 0.400 ## Mean :12.438 Mean : 3.192 ## 3rd Qu.:16.620 3rd Qu.: 2.800 ## Max. :24.470 Max. :46.800 ``` --- # Showing variability: Errorbars ```r ggplot(weather_day, aes(month, TempAve)) + stat_summary(fun=mean, geom="line") + stat_summary(fun.data=mean_se, geom="errorbar") ``` 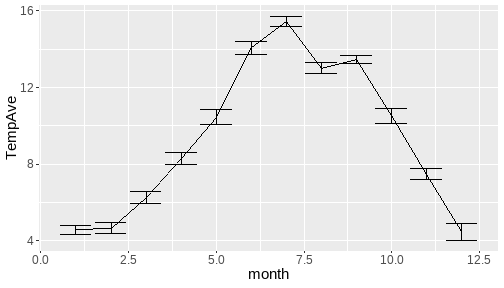<!-- --> --- # Showing variability: Jittered points ```r ggplot(weather_day, aes(month, TempAve)) + geom_jitter(width=0.2) ``` <!-- --> --- # Showing variability: Boxplots ```r ggplot(weather_day, aes(as.factor(month), TempAve)) + geom_boxplot() ``` <!-- --> --- # Showing variability: Violin plots ```r ggplot(weather_day, aes(as.factor(month), TempAve)) + geom_violin() ``` <!-- --> --- # Showing variability: Combinations ```r ggplot(weather_day, aes(as.factor(month), TempAve)) + geom_violin() + geom_jitter(width=0.1, alpha=0.25) + geom_smooth(aes(x=month)) ``` <!-- --> --- # Refining figures * Scales + Colours + Point shapes + Legends/guides + Etc. * Themes + Axes, grid lines, tick marks and labels + Legend position + Font sizes * Annotations: geoms that are not data --- # Example ```r ggplot(weather_day, aes(month, TempAve)) + geom_smooth(color="black") + geom_jitter(width=0.1, alpha=0.5, aes(color=TempAve)) + theme_bw(base_size=12) + scale_color_gradient(low="navy", high="red", guide=FALSE) + labs(x="Month", y="Temperature", title="Edinburgh temperatures in 2014") ``` 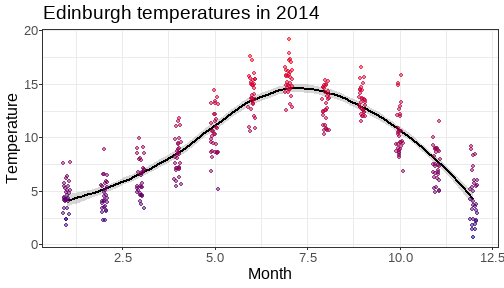<!-- --> # Data management with `tidyverse` Raw data are rarely in a convenient format for analysis. Need to be pre-processed to be ready for analysis. Approx. 80% of analysis time is spent on getting data ready for analysis. ### Basics * Preserve raw data * Save processed data in `.Rdata` files + `save(dat1, dat2, ..., file="FileName.RData")` + Later, you can restore all of those variables using `load("FileName.RData")` * Comment your code --- # Data Transformation: `dplyr` * `filter` rows (observations) * `select` columns (variables) * `arrange` rows * `mutate` create new variables * `summarize` create summaries * `group_by` create grouped summaries (and mutations and filters) * `merge` data sets (base R) * `join` data sets (`dplyr`) --- # Data Transformation: `tidyr` Tidy data > "Happy families are all alike; every unhappy family is unhappy in its own way." - Leo Tolstoy > "Tidy datasets are all alike, but every messy dataset is messy in its own way." - Hadley Wickham  --- # Data Transformation: `tidyr` Tidy data * `pivot_longer` * `pivot_wider` --- # Pipes  Easy way to do a series of operations: `g(f(x, y), z)` is the same as `x %>% f(y) %>% g(z)` But easier to read and write: "Take `x` then do `f(y)` then do `g(z)`" --- # Pipes  (source: https://twitter.com/dmi3k/status/1191824875842879489) --- # Example: Public Health England data on mental health A few interesting and "robust" indicators: * 848 = overall depression prevalence * 90646 = overall depression incidence * 90275 = enough physical activity * 41001 = suicide rate per 100,000 ```r library(tidyverse) ``` --- # Example: Public Health England data on mental health ```r load("./data/PHE_MentalHealth.Rdata") summary(mh_phe) ``` ``` ## IndicatorID IndicatorName County Region ## Min. : 848 Length:9422 Length:9422 Length:9422 ## 1st Qu.:41001 Class :character Class :character Class :character ## Median :41001 Mode :character Mode :character Mode :character ## Mean :44945 ## 3rd Qu.:41001 ## Max. :90646 ## ## Timeperiod Value Year ## Length:9422 Min. : 0.0532 Min. :2001 ## Class :character 1st Qu.: 5.6154 1st Qu.:2006 ## Mode :character Median : 9.5482 Median :2011 ## Mean :12.7162 Mean :2010 ## 3rd Qu.:14.2544 3rd Qu.:2014 ## Max. :71.1897 Max. :2017 ## NA's :619 ``` --- # Example: Public Health England data on mental health ```r str(mh_phe) ``` ``` ## 'data.frame': 9422 obs. of 7 variables: ## $ IndicatorID : int 848 848 848 848 848 848 848 848 848 848 ... ## $ IndicatorName: chr "Depression: Recorded prevalence (aged 18+)" "Depression: Recorded prevalence (aged 18+)" "Depression: Recorded prevalence (aged 18+)" "Depression: Recorded prevalence (aged 18+)" ... ## $ County : chr "Hartlepool" "Middlesbrough" "Redcar and Cleveland" "Stockton-on-Tees" ... ## $ Region : chr "North East" "North East" "North East" "North East" ... ## $ Timeperiod : chr "2013/14" "2013/14" "2013/14" "2013/14" ... ## $ Value : num 5.32 5.74 7.86 9.19 7.38 ... ## $ Year : num 2013 2013 2013 2013 2013 ... ``` --- # Table: Health Indicators by Region ```r *mh_phe %>% group_by(IndicatorName, Region) %>% summarize(Value = mean(Value, na.rm = TRUE)) %>% pivot_wider(names_from = IndicatorName, values_from = Value) ``` --- # Table: Health Indicators by Region ```r mh_phe %>% group_by(IndicatorName, Region) %>% * summarize(Value = mean(Value, na.rm = TRUE)) %>% pivot_wider(names_from = IndicatorName, values_from = Value) ``` --- # Table: Health Indicators by Region ```r mh_phe %>% group_by(IndicatorName, Region) %>% summarize(Value = mean(Value, na.rm = TRUE)) %>% * pivot_wider(names_from = IndicatorName, values_from = Value) ``` ``` ## `summarise()` regrouping output by 'IndicatorName' (override with `.groups` argument) ``` ``` ## # A tibble: 9 x 5 ## Region `Depression: QOF ~ `Depression: Rec~ `Percentage of ph~ `Suicide rate` ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 East M~ 1.49 8.68 56.9 9.90 ## 2 East o~ 1.35 8.03 56.6 9.96 ## 3 London 1.00 6.01 57.3 9.61 ## 4 North ~ 1.47 9.01 52.9 12.4 ## 5 North ~ 1.65 9.69 53.1 11.6 ## 6 South ~ 1.27 8.05 58.7 10.6 ## 7 South ~ 1.33 8.30 58.6 10.9 ## 8 West M~ 1.44 8.73 54.1 10.1 ## 9 Yorksh~ 1.43 8.80 55.2 10.7 ``` --- # Table: Health Indicators by Region .scroll-output[ ```r mh_phe %>% group_by(IndicatorName, Region) %>% summarize(Value = mean(Value, na.rm = TRUE)) %>% pivot_wider(names_from = IndicatorName, values_from = Value) %>% * knitr::kable(digits = 2, format = 'html') ``` ``` ## `summarise()` regrouping output by 'IndicatorName' (override with `.groups` argument) ``` <table> <thead> <tr> <th style="text-align:left;"> Region </th> <th style="text-align:right;"> Depression: QOF incidence (18+) - new diagnosis </th> <th style="text-align:right;"> Depression: Recorded prevalence (aged 18+) </th> <th style="text-align:right;"> Percentage of physically active adults - historical method </th> <th style="text-align:right;"> Suicide rate </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> East Midlands </td> <td style="text-align:right;"> 1.49 </td> <td style="text-align:right;"> 8.68 </td> <td style="text-align:right;"> 56.94 </td> <td style="text-align:right;"> 9.90 </td> </tr> <tr> <td style="text-align:left;"> East of England </td> <td style="text-align:right;"> 1.35 </td> <td style="text-align:right;"> 8.03 </td> <td style="text-align:right;"> 56.58 </td> <td style="text-align:right;"> 9.96 </td> </tr> <tr> <td style="text-align:left;"> London </td> <td style="text-align:right;"> 1.00 </td> <td style="text-align:right;"> 6.01 </td> <td style="text-align:right;"> 57.34 </td> <td style="text-align:right;"> 9.61 </td> </tr> <tr> <td style="text-align:left;"> North East </td> <td style="text-align:right;"> 1.47 </td> <td style="text-align:right;"> 9.01 </td> <td style="text-align:right;"> 52.88 </td> <td style="text-align:right;"> 12.44 </td> </tr> <tr> <td style="text-align:left;"> North West </td> <td style="text-align:right;"> 1.65 </td> <td style="text-align:right;"> 9.69 </td> <td style="text-align:right;"> 53.11 </td> <td style="text-align:right;"> 11.61 </td> </tr> <tr> <td style="text-align:left;"> South East </td> <td style="text-align:right;"> 1.27 </td> <td style="text-align:right;"> 8.05 </td> <td style="text-align:right;"> 58.71 </td> <td style="text-align:right;"> 10.62 </td> </tr> <tr> <td style="text-align:left;"> South West </td> <td style="text-align:right;"> 1.33 </td> <td style="text-align:right;"> 8.30 </td> <td style="text-align:right;"> 58.57 </td> <td style="text-align:right;"> 10.90 </td> </tr> <tr> <td style="text-align:left;"> West Midlands </td> <td style="text-align:right;"> 1.44 </td> <td style="text-align:right;"> 8.73 </td> <td style="text-align:right;"> 54.05 </td> <td style="text-align:right;"> 10.05 </td> </tr> <tr> <td style="text-align:left;"> Yorkshire and the Humber </td> <td style="text-align:right;"> 1.43 </td> <td style="text-align:right;"> 8.80 </td> <td style="text-align:right;"> 55.16 </td> <td style="text-align:right;"> 10.66 </td> </tr> </tbody> </table> ] --- # Table: Health Indicators by Year .scroll-output[ ```r *mh_phe %>% group_by(IndicatorName, Year) %>% summarize(Value = mean(Value, na.rm = TRUE)) %>% pivot_wider(names_from = IndicatorName, values_from = Value) %>% arrange(Year) %>% knitr::kable(digits = 2, format = 'html') ``` ] --- # Table: Health Indicators by Year .scroll-output[ ```r mh_phe %>% group_by(IndicatorName, Year) %>% summarize(Value = mean(Value, na.rm = TRUE)) %>% pivot_wider(names_from = IndicatorName, values_from = Value) %>% * arrange(Year) %>% knitr::kable(digits = 2, format = 'html') ``` ``` ## `summarise()` regrouping output by 'IndicatorName' (override with `.groups` argument) ``` <table> <thead> <tr> <th style="text-align:right;"> Year </th> <th style="text-align:right;"> Depression: QOF incidence (18+) - new diagnosis </th> <th style="text-align:right;"> Depression: Recorded prevalence (aged 18+) </th> <th style="text-align:right;"> Percentage of physically active adults - historical method </th> <th style="text-align:right;"> Suicide rate </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 2001 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 11.25 </td> </tr> <tr> <td style="text-align:right;"> 2002 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 11.01 </td> </tr> <tr> <td style="text-align:right;"> 2003 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.95 </td> </tr> <tr> <td style="text-align:right;"> 2004 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.82 </td> </tr> <tr> <td style="text-align:right;"> 2005 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.39 </td> </tr> <tr> <td style="text-align:right;"> 2006 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.30 </td> </tr> <tr> <td style="text-align:right;"> 2007 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.36 </td> </tr> <tr> <td style="text-align:right;"> 2008 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.26 </td> </tr> <tr> <td style="text-align:right;"> 2009 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.23 </td> </tr> <tr> <td style="text-align:right;"> 2010 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.27 </td> </tr> <tr> <td style="text-align:right;"> 2011 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.60 </td> </tr> <tr> <td style="text-align:right;"> 2012 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 55.67 </td> <td style="text-align:right;"> 10.68 </td> </tr> <tr> <td style="text-align:right;"> 2013 </td> <td style="text-align:right;"> 1.04 </td> <td style="text-align:right;"> 6.44 </td> <td style="text-align:right;"> 55.51 </td> <td style="text-align:right;"> 10.83 </td> </tr> <tr> <td style="text-align:right;"> 2014 </td> <td style="text-align:right;"> 1.18 </td> <td style="text-align:right;"> 7.24 </td> <td style="text-align:right;"> 56.48 </td> <td style="text-align:right;"> 10.62 </td> </tr> <tr> <td style="text-align:right;"> 2015 </td> <td style="text-align:right;"> 1.43 </td> <td style="text-align:right;"> 8.17 </td> <td style="text-align:right;"> 56.54 </td> <td style="text-align:right;"> 10.40 </td> </tr> <tr> <td style="text-align:right;"> 2016 </td> <td style="text-align:right;"> 1.51 </td> <td style="text-align:right;"> 9.04 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> 10.31 </td> </tr> <tr> <td style="text-align:right;"> 2017 </td> <td style="text-align:right;"> 1.55 </td> <td style="text-align:right;"> 9.78 </td> <td style="text-align:right;"> NA </td> <td style="text-align:right;"> NA </td> </tr> </tbody> </table> ] # Reproducible Research * Scientific findings have to be _replicable_. Reproducibility is a pre-requisite for replicability: if I can't reproduce what you did, then I can't check whether it replicates. * Makes your work easier and more efficient + Avoid errors because each step is documented and checkable. + Easier to collaborate because full analysis path is easily shareable. + Easier to update in response to feedback, reviews, etc. + Higher impact: can share your methods as well as your results. _Nature_ journals are moving toward requiring code sharing to increase reproducibility and transparency (e.g., Eglen et al., 2017) --- # Workflow 1. Collect data 2. Read in raw data: `read.table`, `haven::read_sav`, `readxl::read_excel`, `load(url(...))`, etc. 3. Check the data, make exploratory plots: `ggplot` 4. Pre-process the raw data to get it ready for analysis: `filter`, `group_by`, `mutate`, `pivot_longer`, `pivot_wider`, etc. 5. Check the data, make exploratory plots: `ggplot` 6. Analyze the data: `t.test`, `lm`, `lmer`, `sem`, etc. 7. Make final publication-quality figures: `ggplot` 8. Write the report Use R Markdown for steps 2-7 to produce a fully reproducible analysis report. (Can also do step 8 and generate a final PDF or Word document of the full report) --- # R Markdown <img src="./figs/rmarkdown_wizards.png" width="800px" /> --- # R Markdown: Getting Started .pull-left[**Prerequisites** * You'll need the `rmarkdown` and `knitr` packages * Help is readily available: + RStudio --> Help --> Markdown Quick Reference + RStudio --> Help --> Cheatsheets --> R Markdown Cheat Sheet + RStudio --> Help --> Cheatsheets --> R Markdown Reference Guide ] .pull-right[ **Create a markdown file** <img src="./figs/markdown_create.png" width="177px" height="485px" /> ] --- # R Markdown: Template <img src="./figs/markdown_template.png" width="50%" /> --- # R Markdown: Example Excerpt from the R Markdown file for lecture slides: <img src="./figs/markdown_example.png" width="50%" />